v.0.0.7
Launch
Uses GOTTCHA2 to provide taxonomic classifications of shotgun metagenomic reads data.
This is a KBase wrapper for GOTTCHA2, a novel, gene-independent, and signature-based metagenomic taxonomic profiling tool with low false discovery rates (FDR). GOTTCHA2 has been tested and validated on twenty synthetic and mock datasets ranging in community composition and complexity, was applied successfully to data generated from spiked environmental and clinical samples, and robustly demonstrates superior performance compared with other available tools.
GOTTCHA2 takes either a metagenomic reads library or a reads set as input. Parameters can be set for the reference database and for minimum thresholds for the abundance calculations. GOTTCHA2 uses two reference databases for classification, which can be downloaded from the GOTTCHA2 webserver page (last updated 2018-11-10). The databases are:
- RefSeq Bacteria, Archaea and Viruses: Nucleotide unique signature sequences built from NCBI RefSeq Release 90.
- RefSeq Fungal : Fungal Nucleotide unique signature sequences built from NCBI RefSeq Release 90.
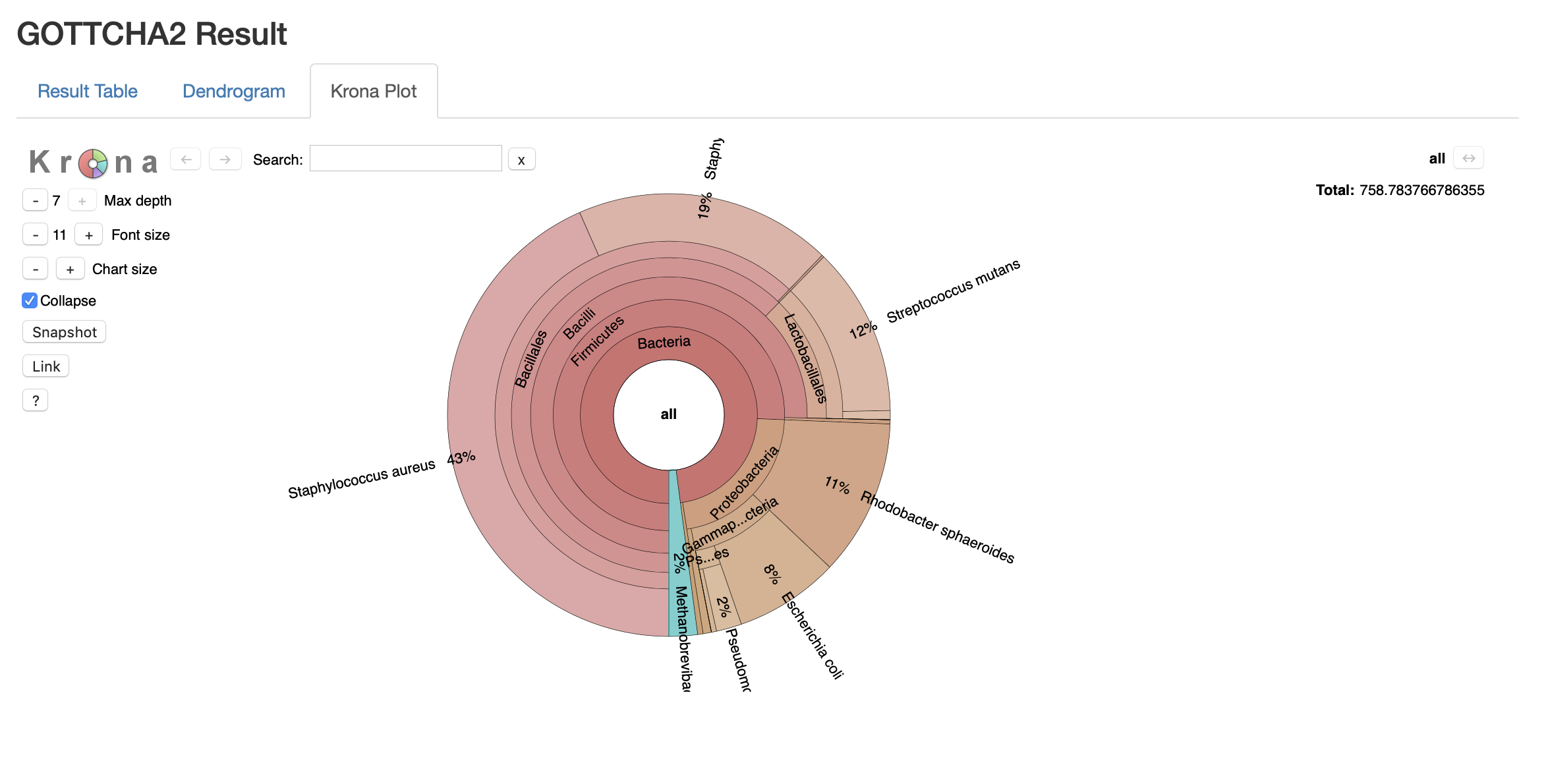
The output of GOTTCHA2 is a report with three modes for analyzing taxonomic abundance. The Result Table lists the taxonomic name and level, relative abundance fraction, and depth of coverage statistics. The Dendrogram sorts the organisms and higher order taxa present into a phylogenetic tree for viewing relationships between organisms within the sample. Finally, the Krona plot is an interactive visualization of the organismal content and relative abundance, allowing the user to focus on abundance within specific clades and generate high quality graphical representations of abundance.
GOTTCHA2 wrapped for KBase by Mark Flynn and Chienchi Lo as part of DOE SFA funded codevelopment with KBase.
Related Publications
- Tracey Allen K. Freitas, Po-E Li, Matthew B. Scholz and Patrick S. G. Chain (2015) Accurate read-based metagenome characterization using a hierarchical suite of unique signatures, Nucleic Acids Research (DOI: 10.1093/nar/gkv180) , http://www.ncbi.nlm.nih.gov/pubmed/25765641
- GOTTCHA2 DBs from: , https://edge-dl.lanl.gov/GOTTCHA2/RefSeq-Release90/
- Krona homepage: , https://github.com/marbl/Krona/wiki
- Github for Krona: , https://github.com/marbl/Krona
App Specification:
https://github.com/LANL-Bioinformatics/kbase-gottcha2.git/tree/f81fc474fdd19568cfbcbf6b0e83339044554f4b/ui/narrative/methods/run_gottcha2Module Commit: f81fc474fdd19568cfbcbf6b0e83339044554f4b