Obtain objective taxonomic assignments for bacterial and archaeal genomes based on the Genome Taxonomy Database (GTDB)
Description
GTDB-Tk v2.3.2 is a software toolkit for assigning objective taxonomic classifications to bacterial and archaeal genomes. It is designed to work with recent advances that allow hundreds or thousands of metagenome-assembled genomes (MAGs) to be obtained directly from environmental samples. It can also be applied to isolate and single-cell genomes. If the input is a GenomeSet, the App will optionally overwrite the Taxonomy information in the Genome object(s).
Notes:As of v2.0.0, the GTDB-Tk Classify workflow by default uses class-level subtrees rather than the entire Bacterial tree. This uses far less memory. An option to use the full tree is provided.
As of v2.0.0, intermediate files in the classification workflow are deleted. An option to keep those intermediate files is provided.
Individual Genome objects must first be placed into a GenomeSet to be used as input. This is to avoid accidentally running the App multiple times, once for each Genome, which is very inefficient. Similarly, individual Assembly objects must be placed into an AssemblySet object. The utility Apps Build GenomeSet or Build AssemblySet can be used for this purpose.
Tool and Data Sources:
GTDB-Tk v2.3.2 is installed from https://github.com/Ecogenomics/GTDBTk
This app is using gtdbtk release 207 data gtdbtk_r207_v2_data.tar.gz and release 214 data gtdbtk_r214_data.tar.gz and accompanying metadata ar53_metadata_r[207/214].tsv and bac120_metadata_r[207/214]. These reference data correspond to the GTDB R07-RS207 and R08-RS214 releases respectively.
The Genome Taxonomy Database (GTDB) is constructed from RefSeq and Genbank genomes, and releases are indexed to RefSeq releases. All genomes are quality controlled using CheckM and those statistics can be found on the GTDB website.
Usage
GTDB-Tk Classify will place your Binned Contigs, Assemblies, or Genomes (from a GenomeSet or a Species Tree) into the GTDB Taxonomy. Additionally, if your queries are Genome objects, the KBase implementation will allow the user to copy the GTDB species representative Genome objects to their Narrative, as well as any members in the Species Tree from the GTDB-Tk classify placement. Options for those copy operations, as well as whether to overwrite any existing taxonomic classification in the query genome objects, are available, as described in the Parameters section below.
In addition to the visual reports (see below) and the GTDB species representative Genome objects, this App has the option of generative KBase Species Tree objects, which can be used in subsequent analyses such as Pangenome Calculations with the mOTUpan App.
Report
GTDB Species Tree Tab
dendrogram / ultrametric tree
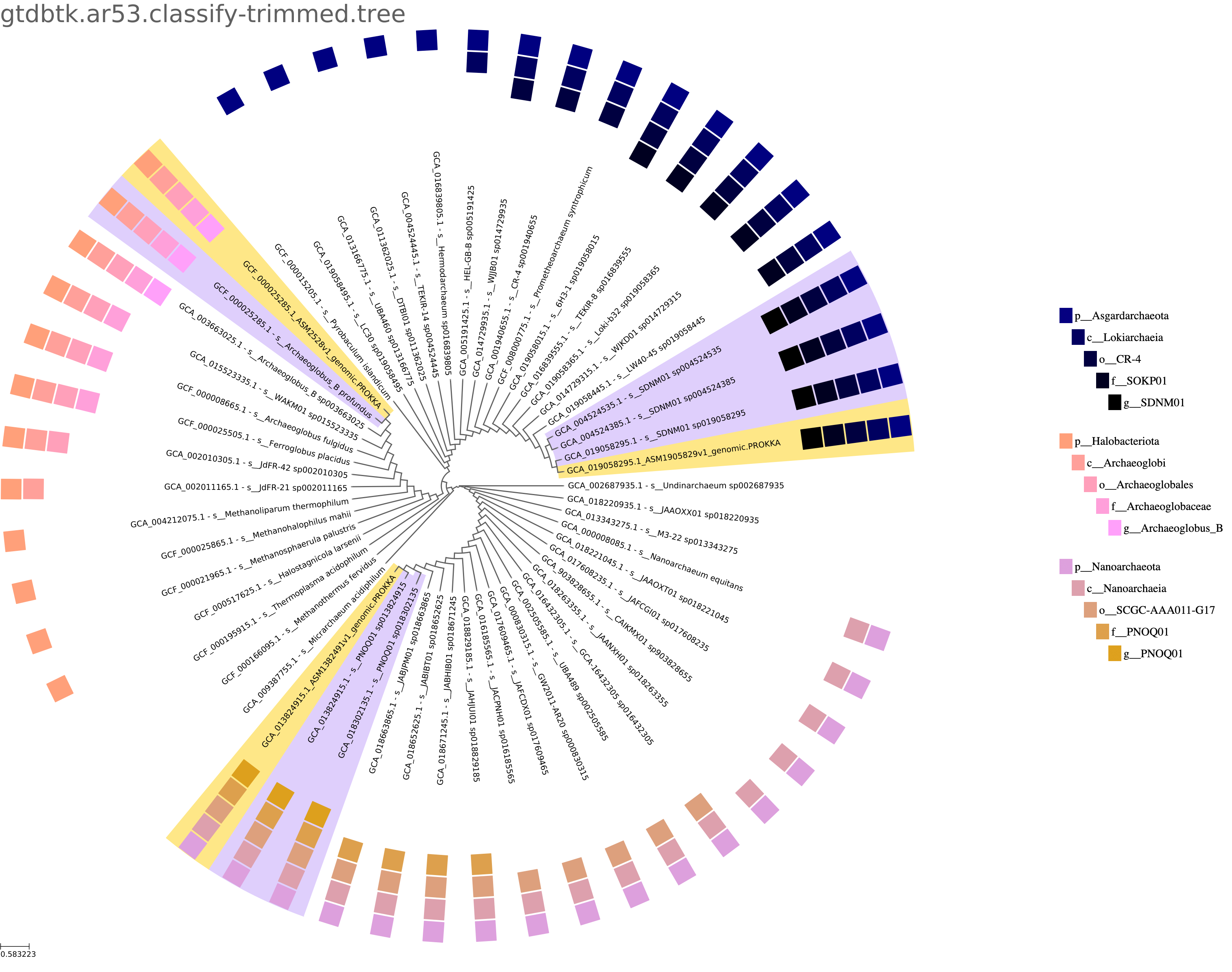
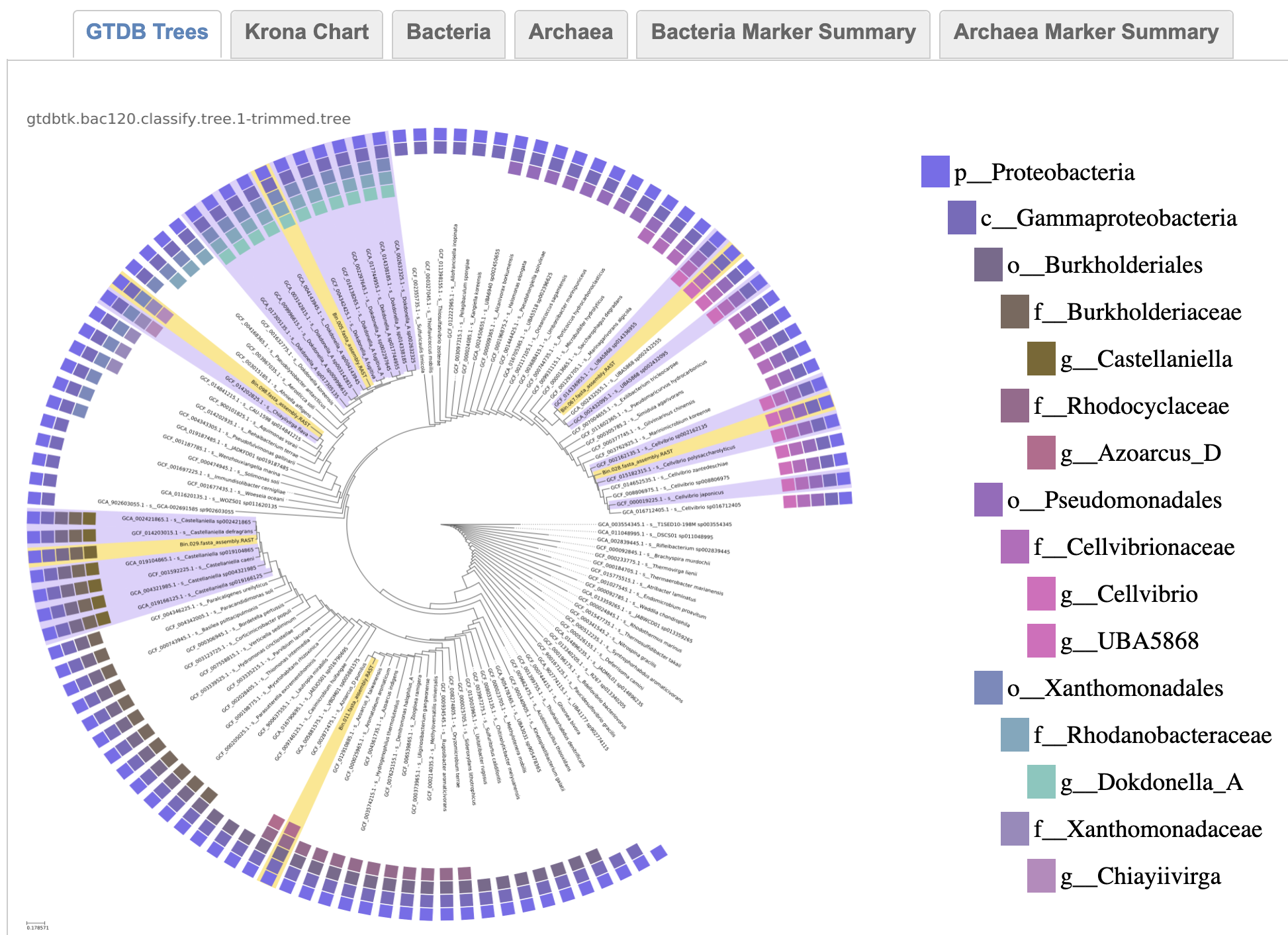
Queries are placed into the GTDB Species Tree. While a uniform dendrogram and a evolutionary distance-based branch length version of the tree are generated by the App, the User has the option of the report including the dendrogram or the phylogenetic branch length version of the image. Both versions are available for Download in both bitmap (PNG) and vector (PDF) formats. A key which indicates taxonomic membership of each leaf is provided corresponding to the outer circle on the plot. One tree for any Archaea and typically one tree per Bacterial phylum are included in the output. The Queries are in yellow and proximal lineages found by GTDB-Tk Classify in lavender. The remaining tree is trimmed to include one species representative per branch attached to the primary branches within which the queries and proximal species are found. These genomes are selected following the same weighting as used by GTDB to select species representatives, with an added term with the greatest weight to select the GTDB genus representatives.
phylogenetic tree
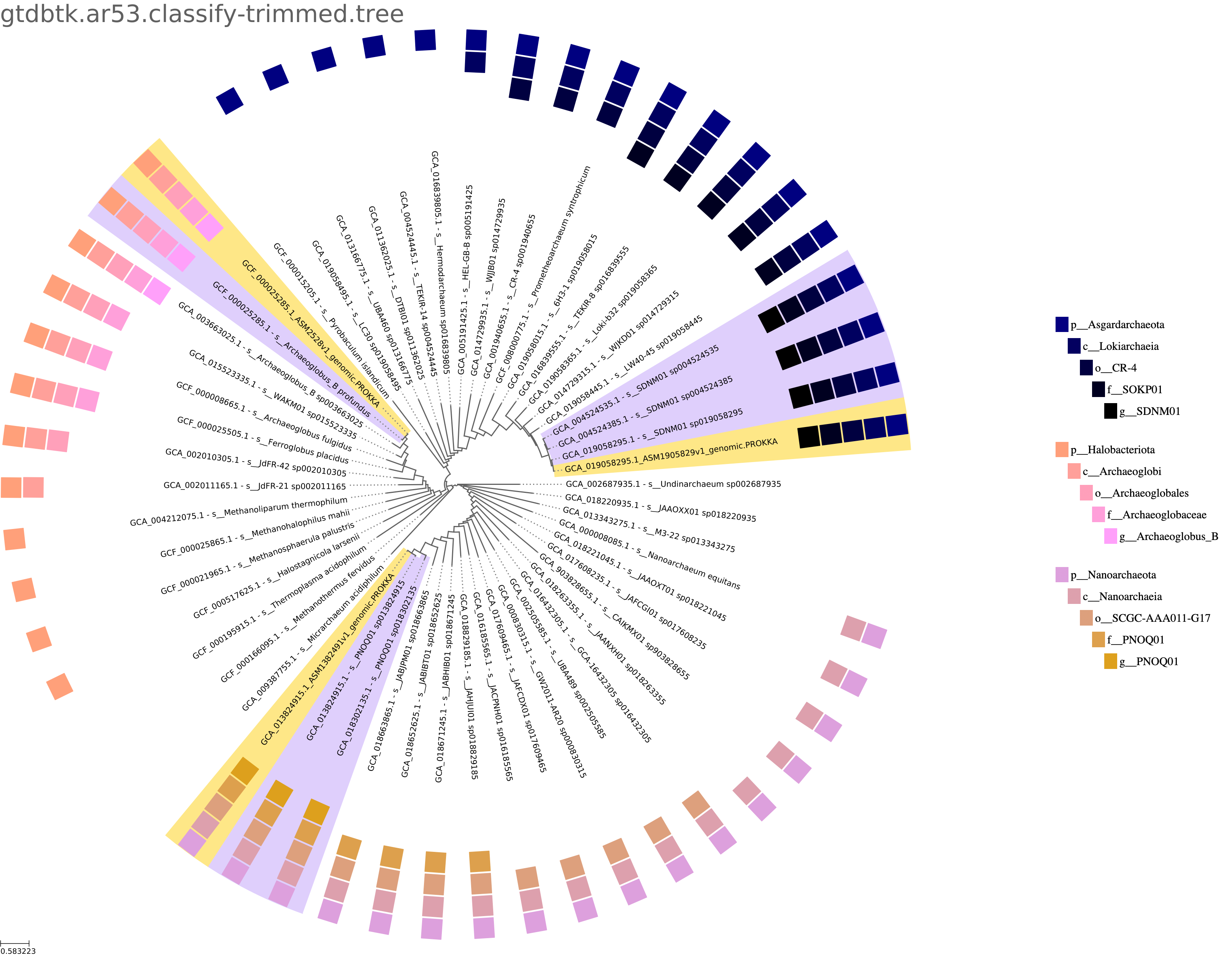
As an alternative to the uniform dendrogram version of the tree, a proper phylogenetic tree with branch lengths indicating evolutionary distance is available and can be configured to be what is shown in the report. Regardless, it can be downloaded as the tree without the label "ultrametric".
rectangular tree
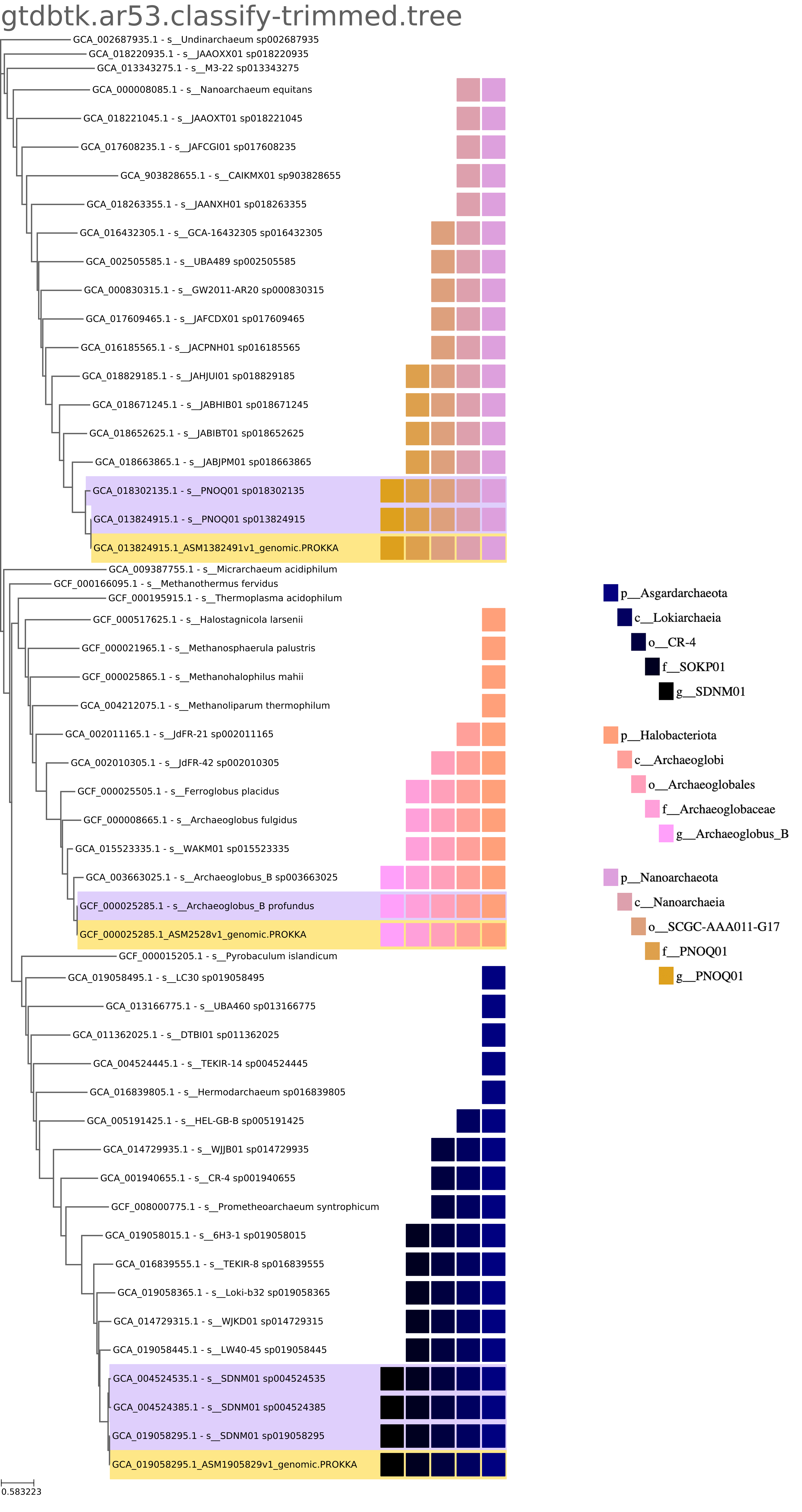
Rectangular tree images are also generated and available for download.
bacterial trees
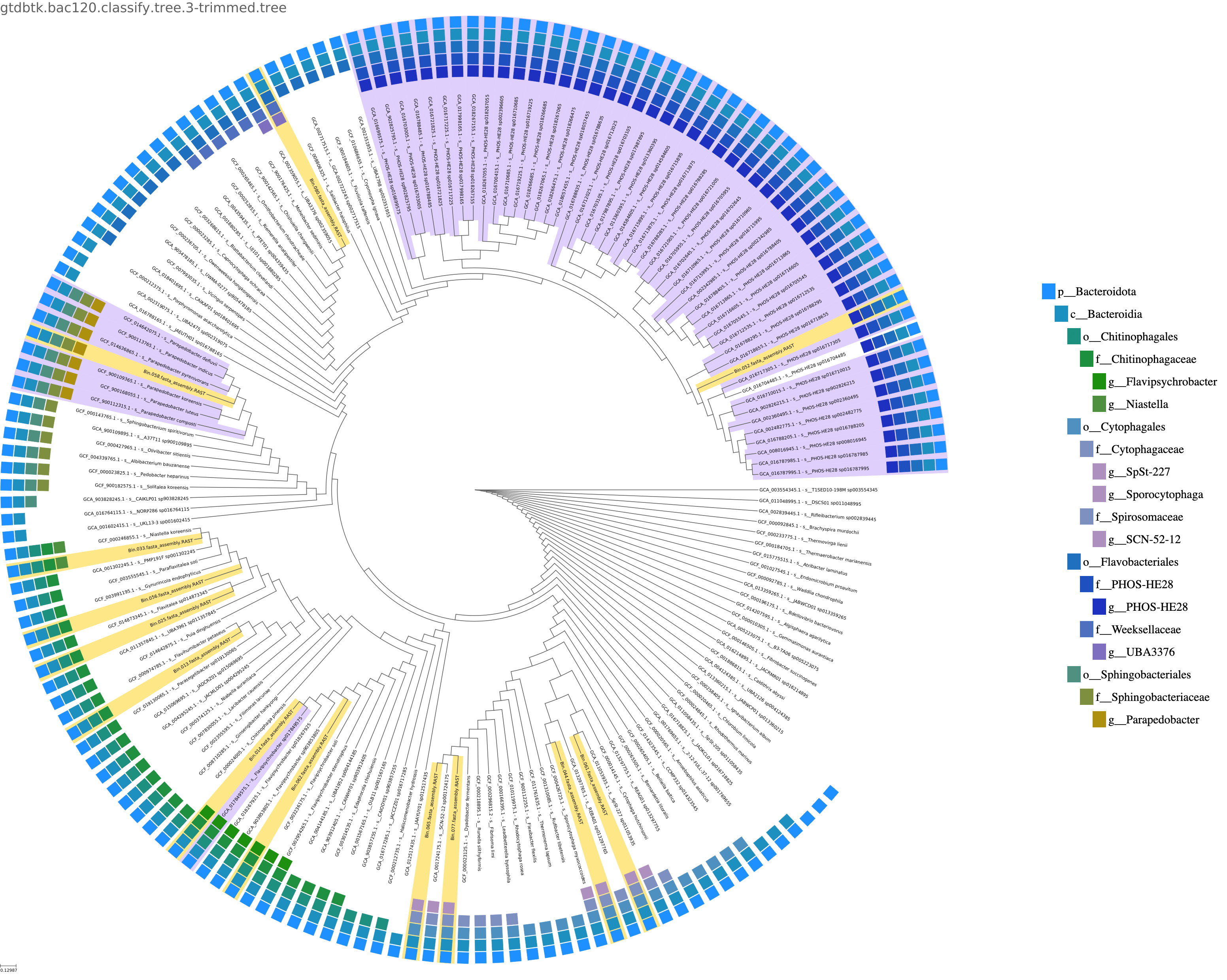
While the Archaea are all placed within a single tree, Bacterial queries are placed within separate trees, typically one phylum per tree. There is also often a "backbone" tree at a higher taxonomic level where queries that don't fall within class trees can be found.
Krona Plot Tab
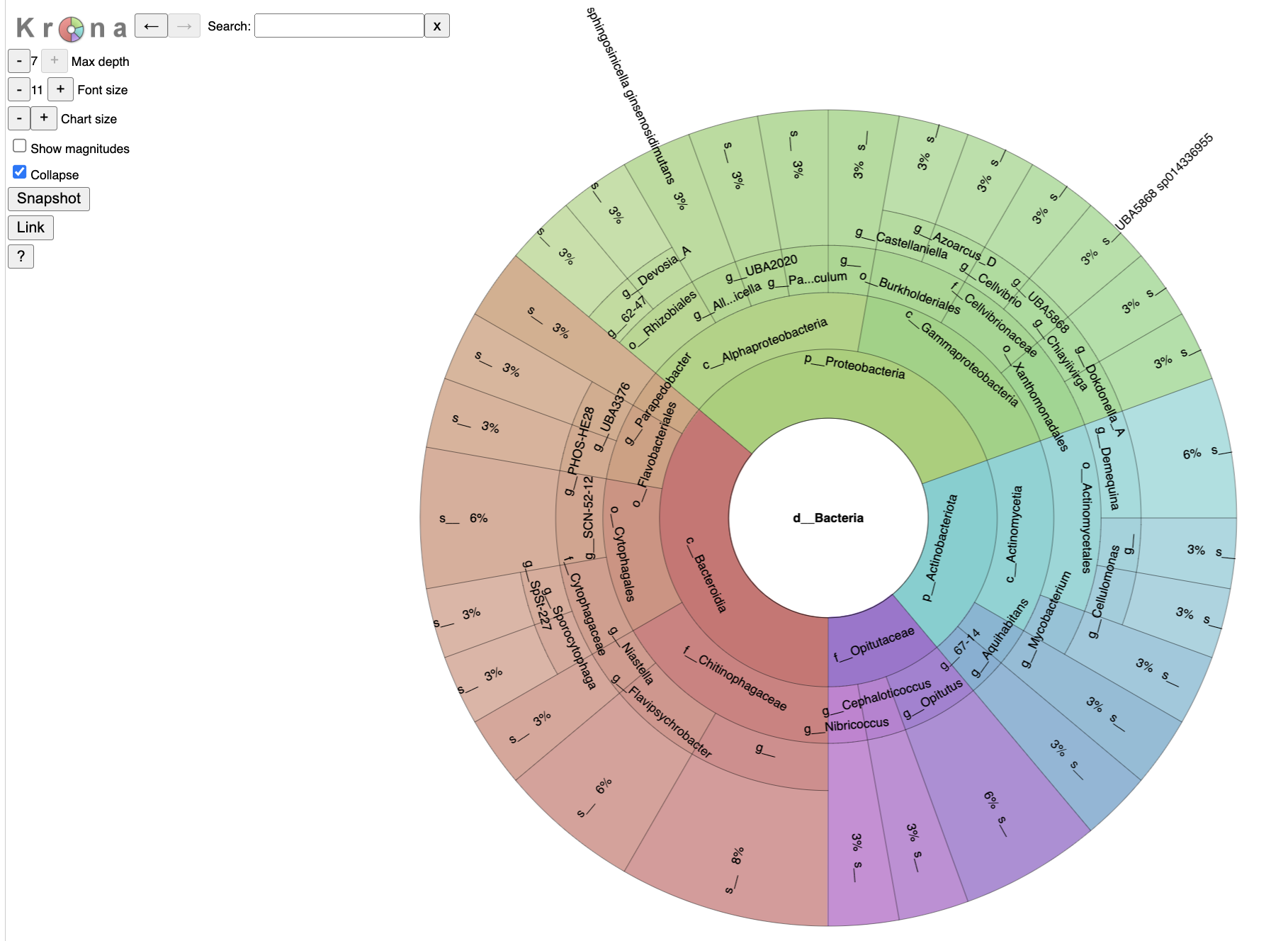
The classifications will also be available using the Krona visualization.
Bacteria and Archaea Classification Tabs
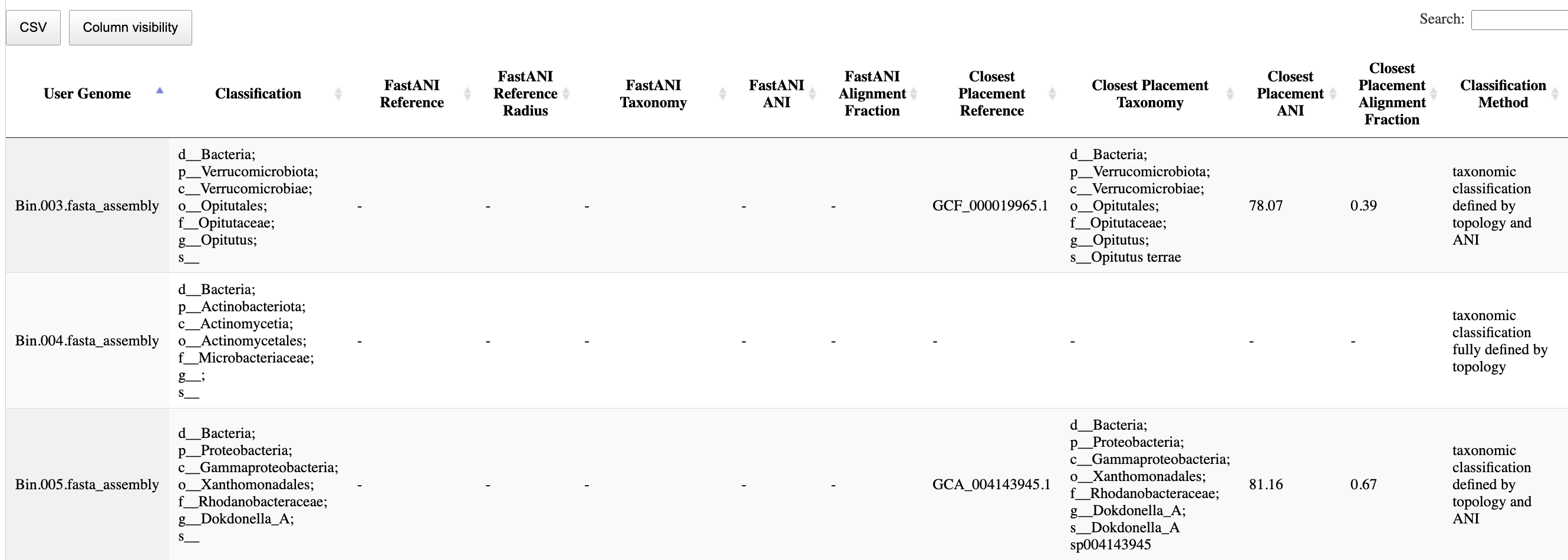
A table with the classifications is also available in the App report. Bacterial and Archaeal classifications are in separate tabs. Columns for the consensus classification (often only to a higher taxonomic level than species) and ANI assignment are given.
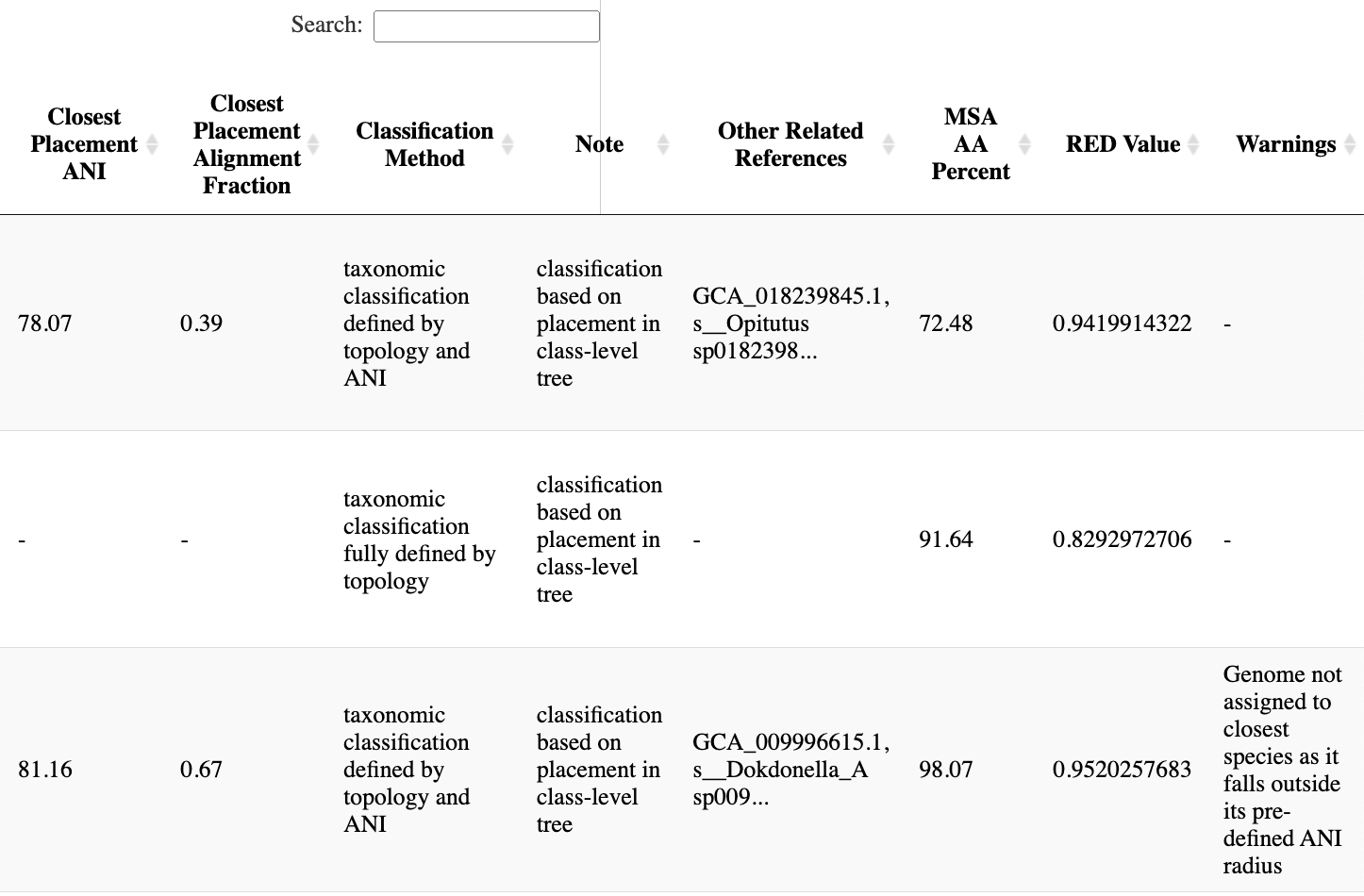
The classification tabs also include tree-placement-based classification, terms used in the determination of the placement, and other notes and the GTDB ID of additional proximal species representative genomes. Please see the GTDB-Tk paper for the full details.
Bacteria and Archaea Marker Gene Tabs
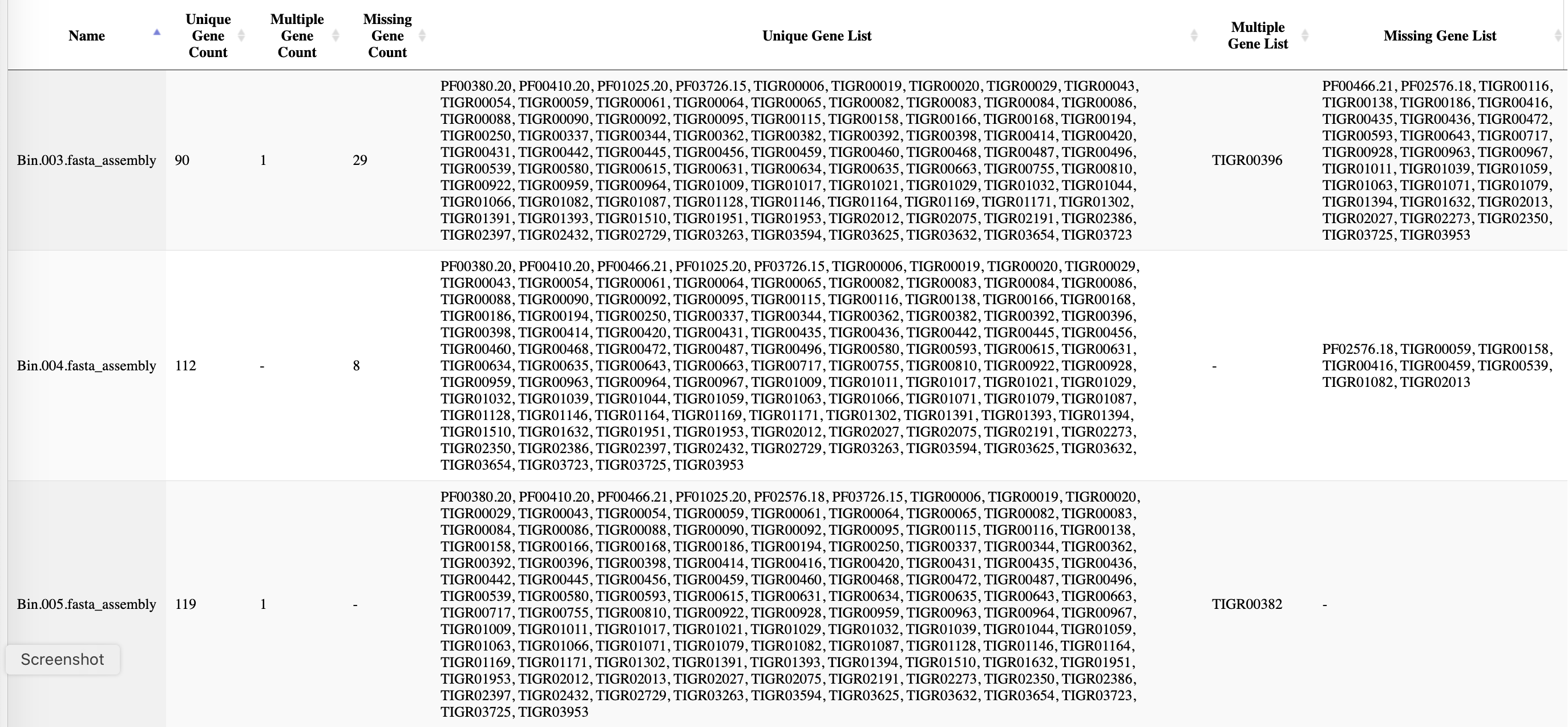
Lastly there are two tabs, one for Bacteria and one for Archaea, with the phylogenetic marker genes used in the taxonomic placement by GTDB-Tk. Often, not all markers are found in a given query, and the presence or absence of a marker is indicated in this table.
Generated objects
If the query objects are Genomes (or refer to Genome objects such as a Species Tree query), then the user has the choice of generating the following output objects from the GTDB-Tk Classify App.
Genome objects
The user has the option of copying the GTDB species representative Genome objects into the current workspace. This can be done for the proximal species representatives as well as the more distal lineages found in the trimmed Species Tree (see above). If this is done, any Genome Sets and Species Tree objects will refer to the local Genome copies. Therefore, subsequent analyses, such as Pangenome calculations, will use these local copies.
Genome Sets
Genome Sets are produced for the proximal species representatives identified by GTDB-Tk Classify. The User may wish to adjust the membership of which genomes are in the Genome Set and can do so with Apps such as Remove Genome from GenomeSet or Trim Species Tree to GenomeSet prior to further analysis.
Species Trees
Species Tree objects can also be produced by GTDB-Tk Classify. One will be generated that contains just the query genomes and the proximal species hits, as well as a Species Tree that includes the greater set of distal genomes from the trimmed tree. There will be one for Archaea and one for each Bacterial phylum.
Downloadable Files

The entire working directory used in the GTDB-Tk Classify calculations and all output are availble for download as a zip archive with the file name "GTDB-Tk_classify_wf.zip". Additionally, several key output files generated by the KBase App version of GTDB-Tk Classify can be downloaded, including the PNG and PDF versions of the trees described above, the newick versions of the proximal and trimmed trees, as well as a file with the GTDB lineage for each query and each member of the species trees. This set of downloadable files is available for the Archaea with the prefix "gtdbtk.ar53.classify" and for Bacteria, one per phylum, with the prefix "gtdbtk.bac120.classify.tree.#" where "#" is just an index for the phyla.
Team members who implemented App in KBase: Dylan Chivian and Paramvir Dehal. For questions, please contact us.
If you use GTDB-Tk in KBase, please cite
- Chivian D, Jungbluth SP, Dehal PS, Wood-Charlson EM, Canon RS, Allen BH, Clark MM, Gu T, Land ML, Price GA, Riehl WJ, Sneddon MW, Sutormin R, Zhang Q, Cottingham RW, Henry CS, Arkin AP. Metagenome-assembled genome extraction and analysis from microbiomes using KBase. Nat Protoc. 2023 Jan;18(1):208-238. doi: 10.1038/s41596-022-00747-x
GTDB-Tk is described in:
- Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2022 GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics, btac672
The Genome Taxonomy Database (GTDB) is described in:
- Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil PA, Hugenholtz P. 2022 GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Research, gkab776
We also strongly encourage you to cite the following 3rd party dependencies:
- Matsen FA, Kodner RB, Armbrust EV. 2010. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics, 11:538.
- Jain C, et al. 2017. High-throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. bioRxiv, https://doi.org/10.1101/225342.
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11:119. doi: 10.1186/1471-2105-11-119.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 - Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One, 5, e9490.
- Eddy SR. 2011. Accelerated profile HMM searches. PLOS Comp. Biol., 7:e1002195.
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016 Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17(1):132.
Related Publications
- Pierre-Alain Chaumeil, Aaron J Mussig, Philip Hugenholtz, Donovan H Parks. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics, Volume 38, Issue 23, 1 December 2022, Pages 5315 5316. DOI: https://doi.org/10.1093/bioinformatics/btac672 , https://academic.oup.com/bioinformatics/article/38/23/5315/6758240
- Pierre-Alain Chaumeil, Aaron J Mussig, Philip Hugenholtz, Donovan H Parks, GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database, Bioinformatics, Volume 36, Issue 6, 15 March 2020, Pages 1925 1927. DOI: https://doi.org/10.1093/bioinformatics/btz848 , https://doi.org/10.1093/bioinformatics/btz848
- Donovan H Parks, Maria Chuvochina, Christian Rinke, Aaron J Mussig, Pierre-Alain Chaumeil, Philip Hugenholtz. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Research, Volume 50, Issue D1, 7 January 2022, Pages D785 D794. DOI: https://doi.org/10.1093/nar/gkab776 , https://academic.oup.com/nar/article/50/D1/D785/6370255
- Parks, D., Chuvochina, M., Waite, D. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36, 996 1004 (2018). DOI: https://doi.org/10.1038/nbt.4229 , http://dx.doi.org/10.1038/nbt.4229
- Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, Hugenholtz P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020;10.1038/s41587-020-0501-8. DOI:10.1038/s41587-020-0501-8 , https://www.nature.com/articles/s41587-020-0501-8
- Rinke C, Chuvochina M, Mussig AJ, Chaumeil PA, Dav n AA, Waite DW, Whitman WB, Parks DH, and Hugenholtz P. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat Microbiol. 2021 Jul;6(7):946-959. DOI:10.1038/s41564-021-00918-8 , https://www.nature.com/articles/s41564-021-00918-8
- Chivian D, Jungbluth SP, Dehal PS, Wood-Charlson EM, Canon RS, Allen BH, Clark MM, Gu T, Land ML, Price GA, Riehl WJ, Sneddon MW, Sutormin R, Zhang Q, Cottingham RW, Henry CS, Arkin AP. Metagenome-assembled genome extraction and analysis from microbiomes using KBase. Nat Protoc. 2023 Jan;18(1):208-238. doi: 10.1038/s41596-022-00747-x , https://www.nature.com/articles/s41596-022-00747-x
- Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. Published 2010 Oct 30. doi:10.1186/1471-2105-11-538 , https://pubmed.ncbi.nlm.nih.gov/21034504/
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114. Published 2018 Nov 30. DOI:10.1038/s41467-018-07641-9 , https://www.nature.com/articles/s41467-018-07641-9
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. Published 2010 Mar 8. DOI:10.1186/1471-2105-11-119 , https://pubmed.ncbi.nlm.nih.gov/20211023/
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. Published 2010 Mar 10. DOI:10.1371/journal.pone.0009490 link: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2835736/ ,
- Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7(10):e1002195. DOI:10.1371/journal.pcbi.1002195 , https://pubmed.ncbi.nlm.nih.gov/22039361/
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016 Jun 20;17(1):132. DOI: 10.1186/s13059-016-0997-x , https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-0997-x
App Specification:
https://github.com/kbaseapps/kb_gtdbtk/tree/99839a03cebee1110c6cd6d807dc448821837580/ui/narrative/methods/run_kb_gtdbtk_classify_wfModule Commit: 99839a03cebee1110c6cd6d807dc448821837580