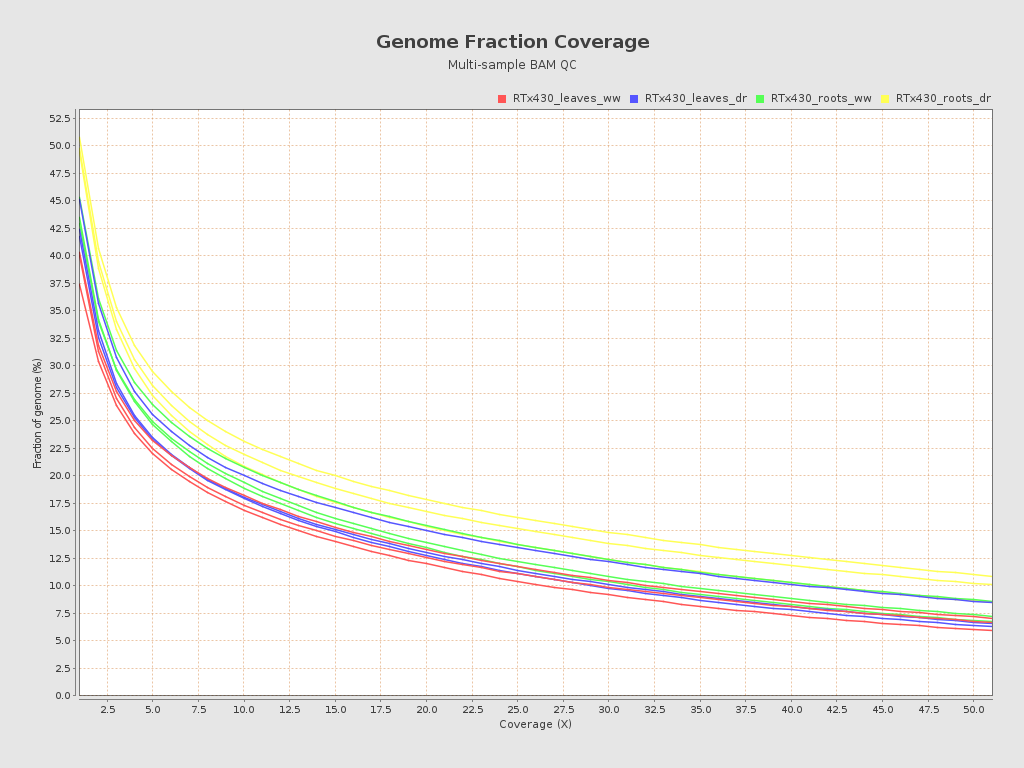
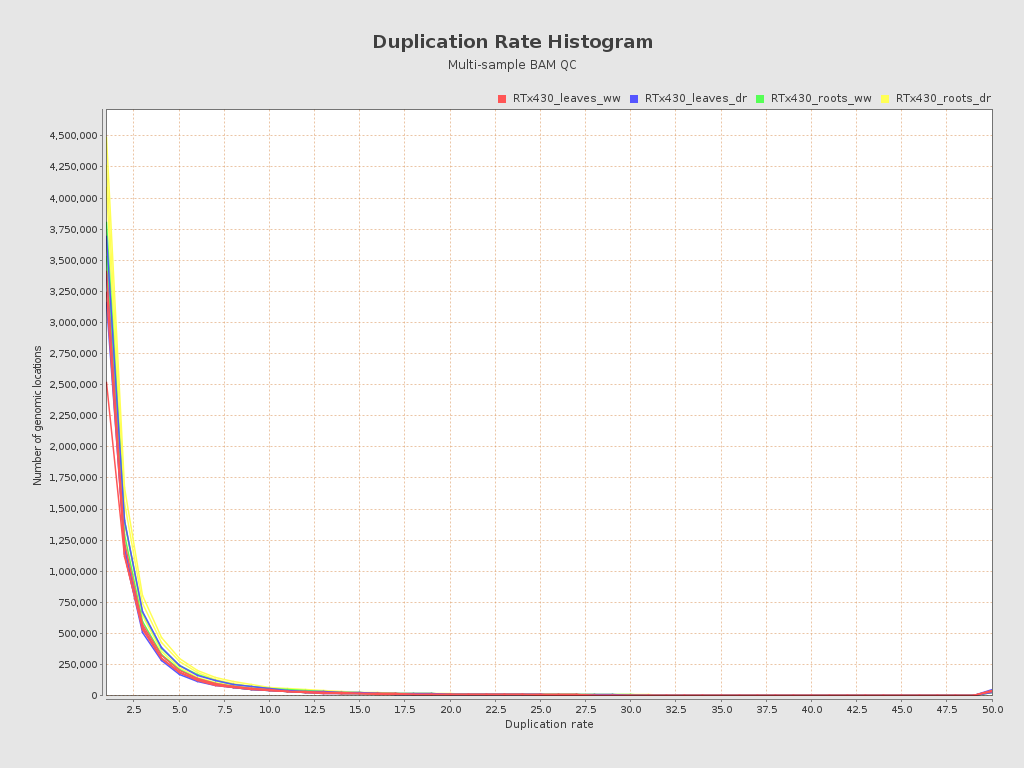
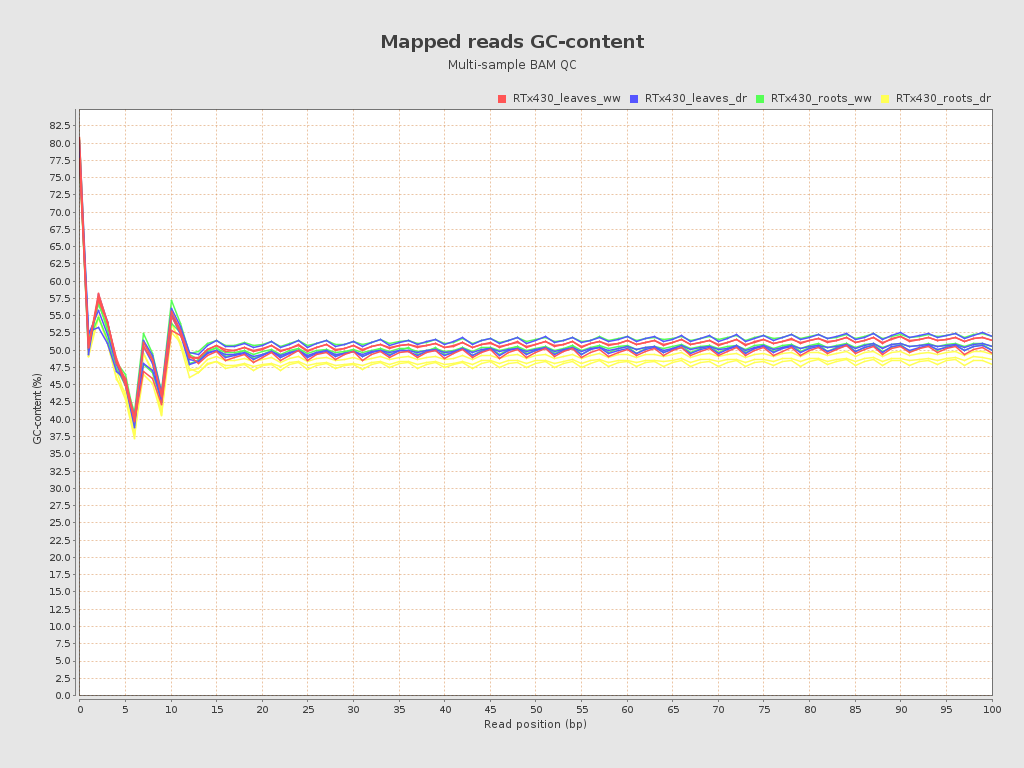
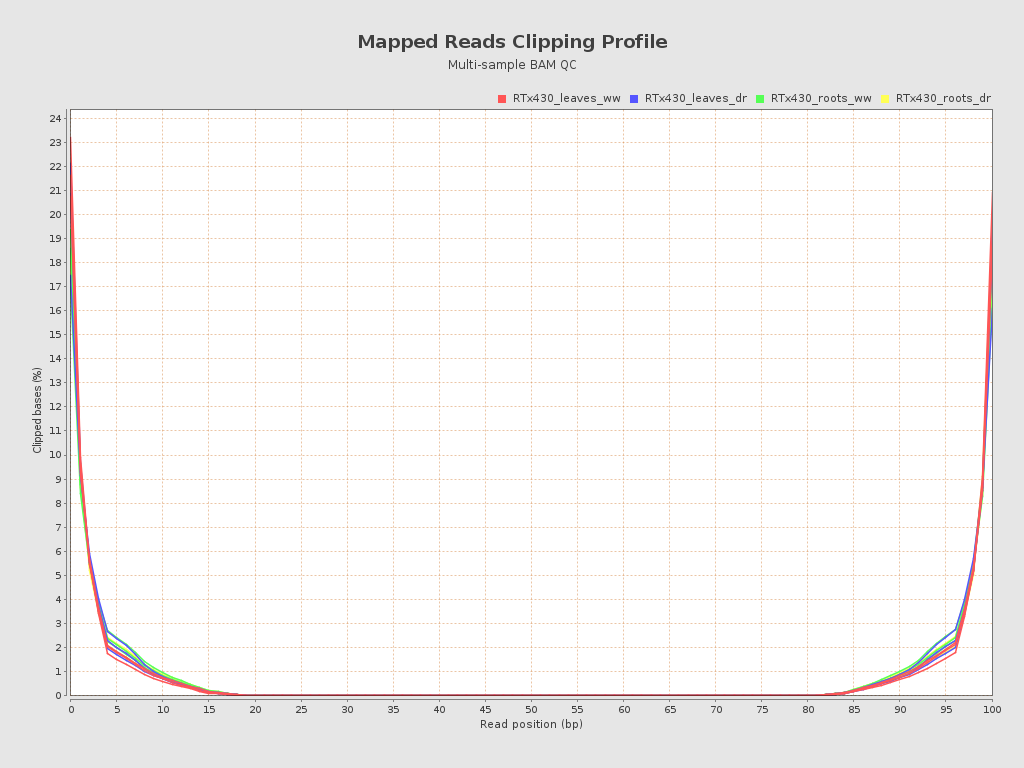
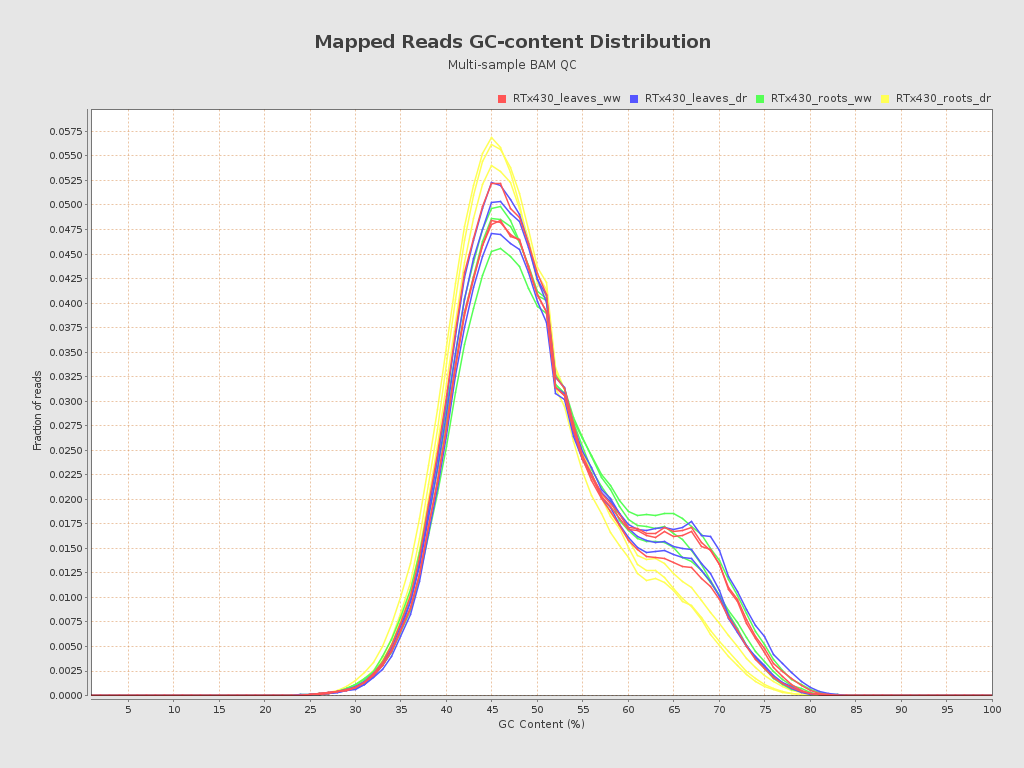
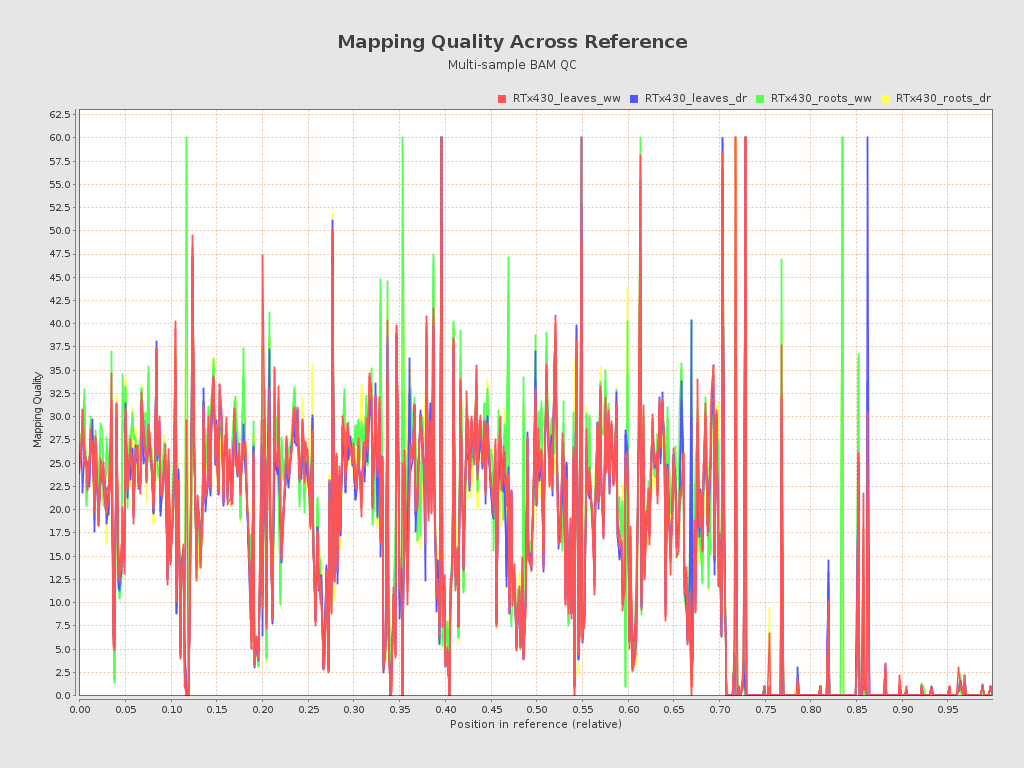
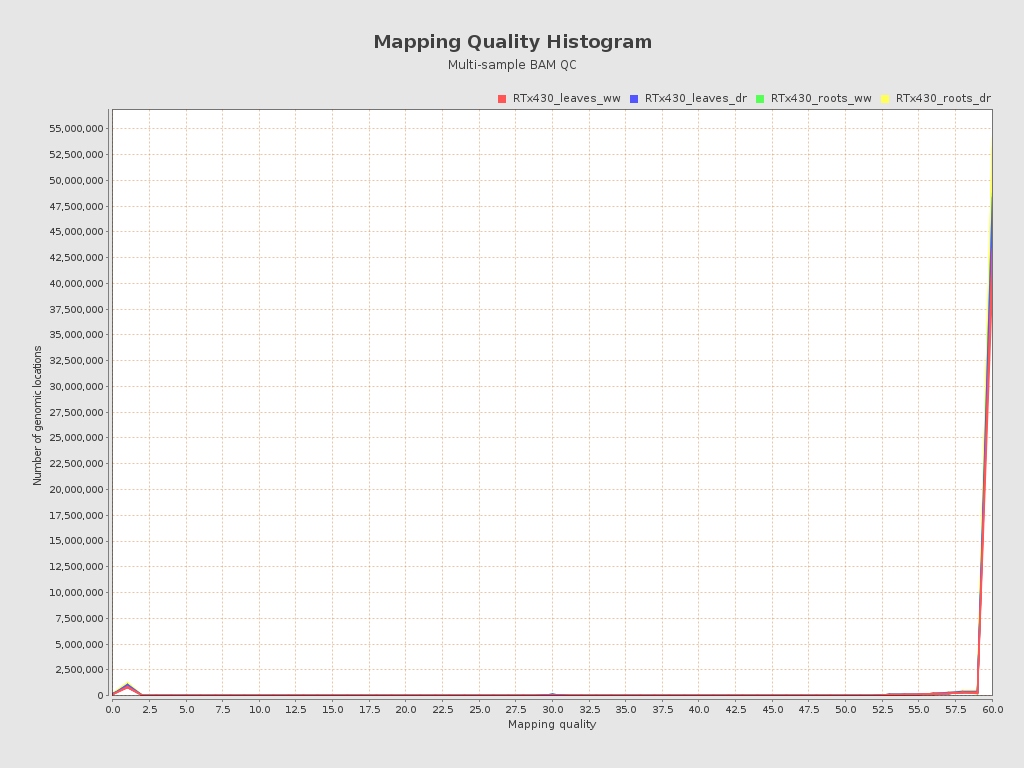
| RTx430_leaves_dr_r1_alignment (RTx430_leaves_dr) |
/kb/module/work/tmp/download_ccb06cd9-2e96-47ff-83eb-6645eb0241fb/accepted_hits_stats |
| RTx430_leaves_ww_r2_alignment (RTx430_leaves_ww) |
/kb/module/work/tmp/download_595f4c76-70b3-404b-89cc-40e5efb11593/accepted_hits_stats |
| RTx430_leaves_dr_r2_alignment (RTx430_leaves_dr) |
/kb/module/work/tmp/download_7028c533-4455-4fa9-8040-8453ada5ef60/accepted_hits_stats |
| RTx430_roots_dr_r2_alignment (RTx430_roots_dr) |
/kb/module/work/tmp/download_20d8780c-5478-496f-a106-3829c8ac0acf/accepted_hits_stats |
| RTx430_roots_dr_r3_alignment (RTx430_roots_dr) |
/kb/module/work/tmp/download_ee6d965a-a56b-4d9e-b875-0031d81055ef/accepted_hits_stats |
| RTx430_leaves_dr_r3_alignment (RTx430_leaves_dr) |
/kb/module/work/tmp/download_f542f2ac-59f4-4b9f-86a4-6cae7753852f/accepted_hits_stats |
| RTx430_roots_ww_r1_alignment (RTx430_roots_ww) |
/kb/module/work/tmp/download_008f5835-4252-438a-bcea-e5b9d9835a56/accepted_hits_stats |
| RTx430_roots_dr_r1_alignment (RTx430_roots_dr) |
/kb/module/work/tmp/download_c8f2715a-8e91-4860-a205-8da02d857fdd/accepted_hits_stats |
| RTx430_leaves_ww_r3_alignment (RTx430_leaves_ww) |
/kb/module/work/tmp/download_1ccbebfd-4849-4f50-ba29-071042d0cf2e/accepted_hits_stats |
| RTx430_roots_ww_r2_alignment (RTx430_roots_ww) |
/kb/module/work/tmp/download_0f03a9a6-8747-4ab8-a77c-32bcd570d9a4/accepted_hits_stats |
| RTx430_roots_ww_r3_alignment (RTx430_roots_ww) |
/kb/module/work/tmp/download_d1517e40-bc3b-4aa9-af20-64a63455a99e/accepted_hits_stats |
| RTx430_leaves_ww_r1_alignment (RTx430_leaves_ww) |
/kb/module/work/tmp/download_c04375d1-784e-4168-880e-8ba35f71322a/accepted_hits_stats |

.png)