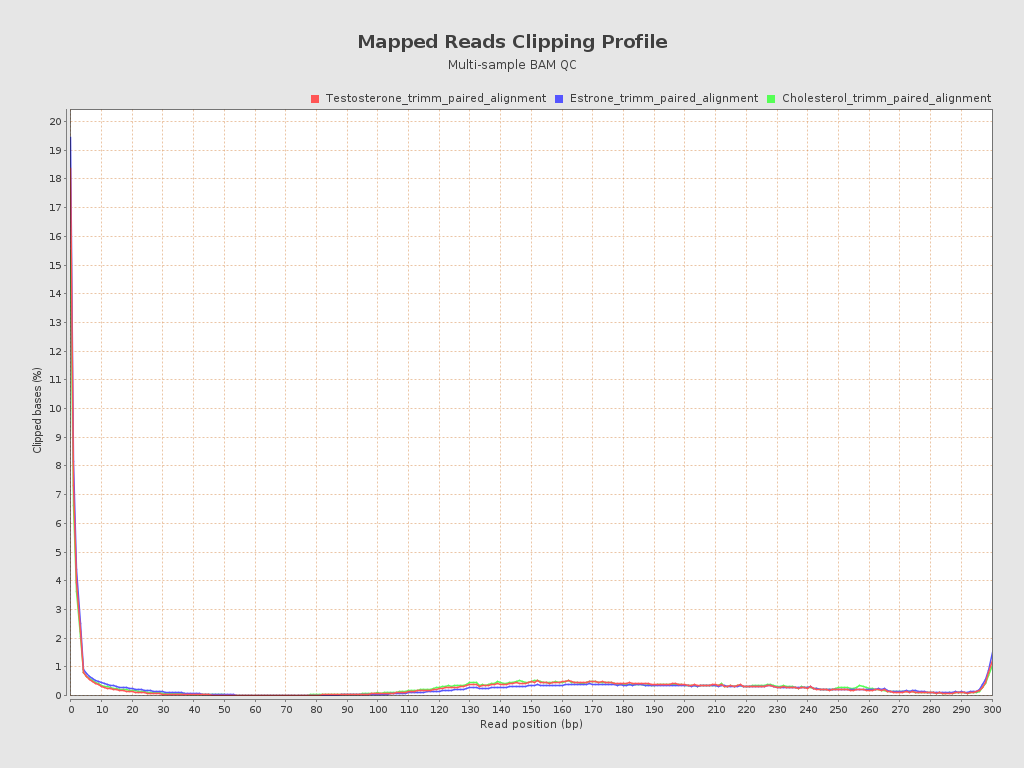
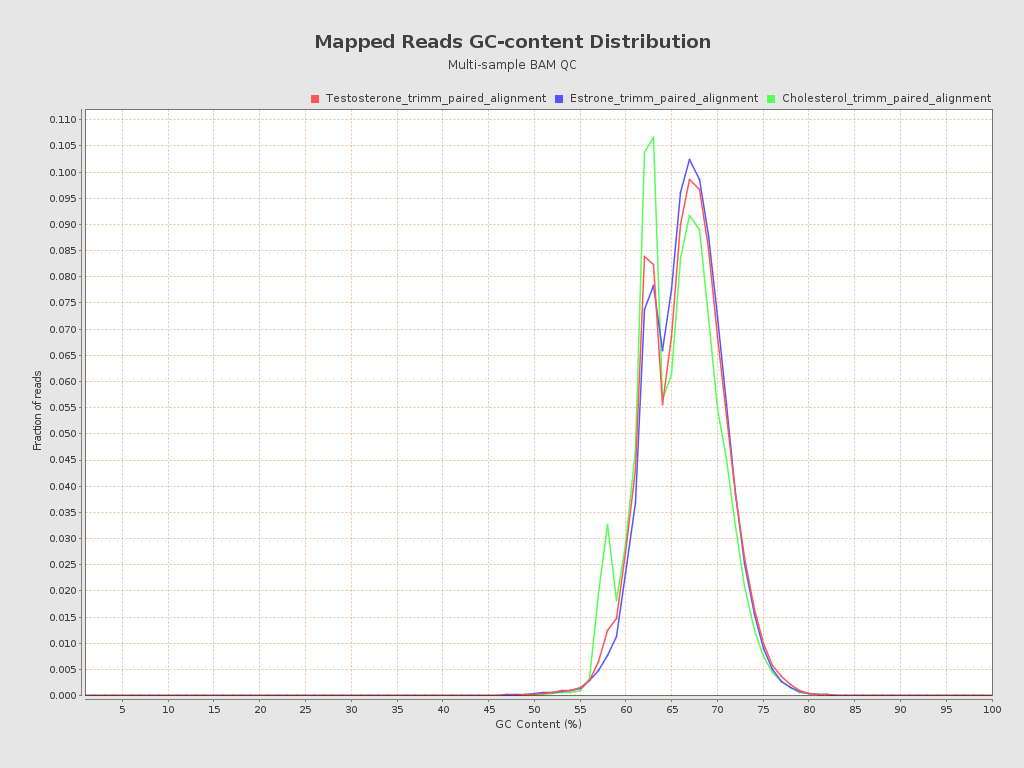
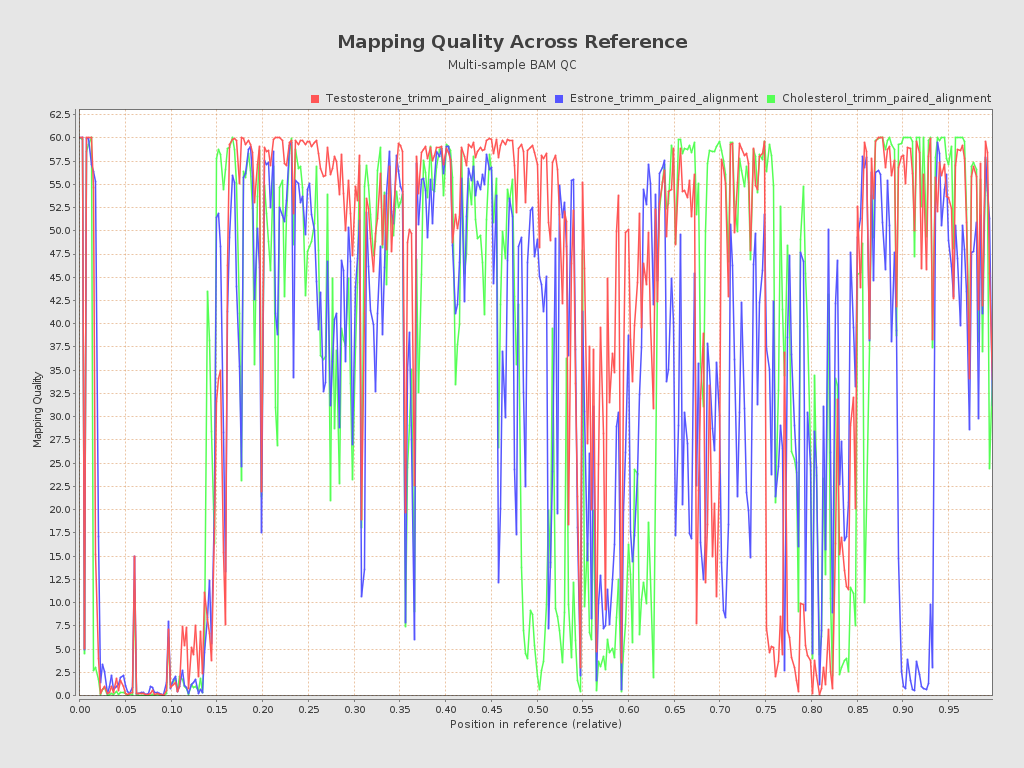
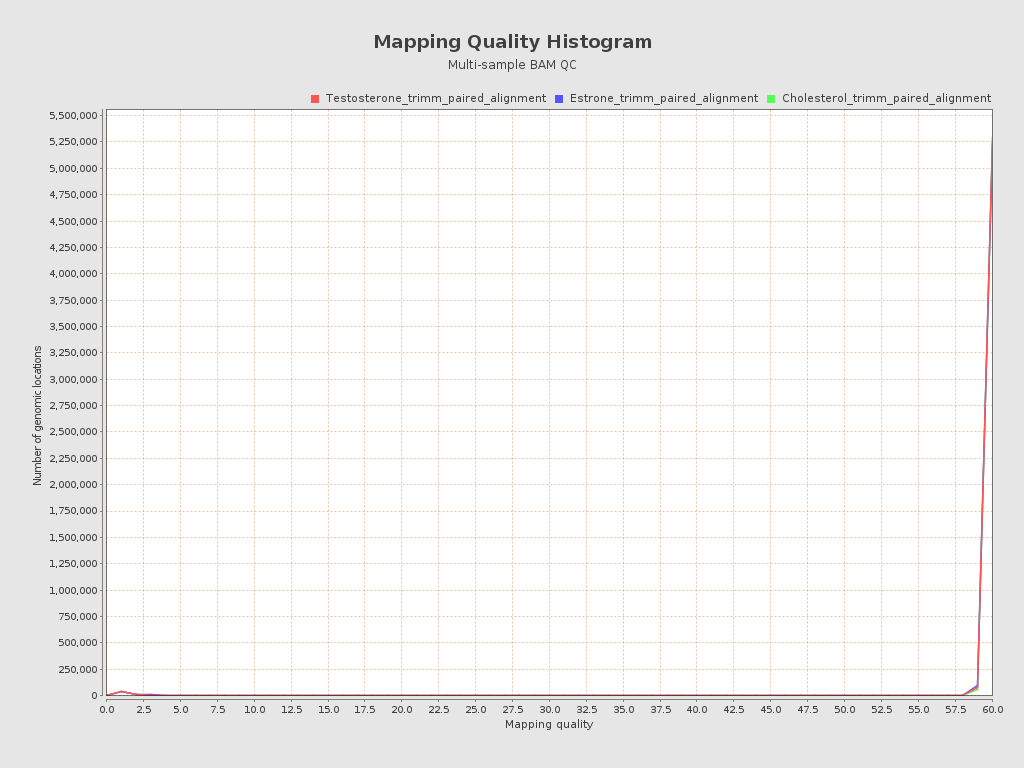
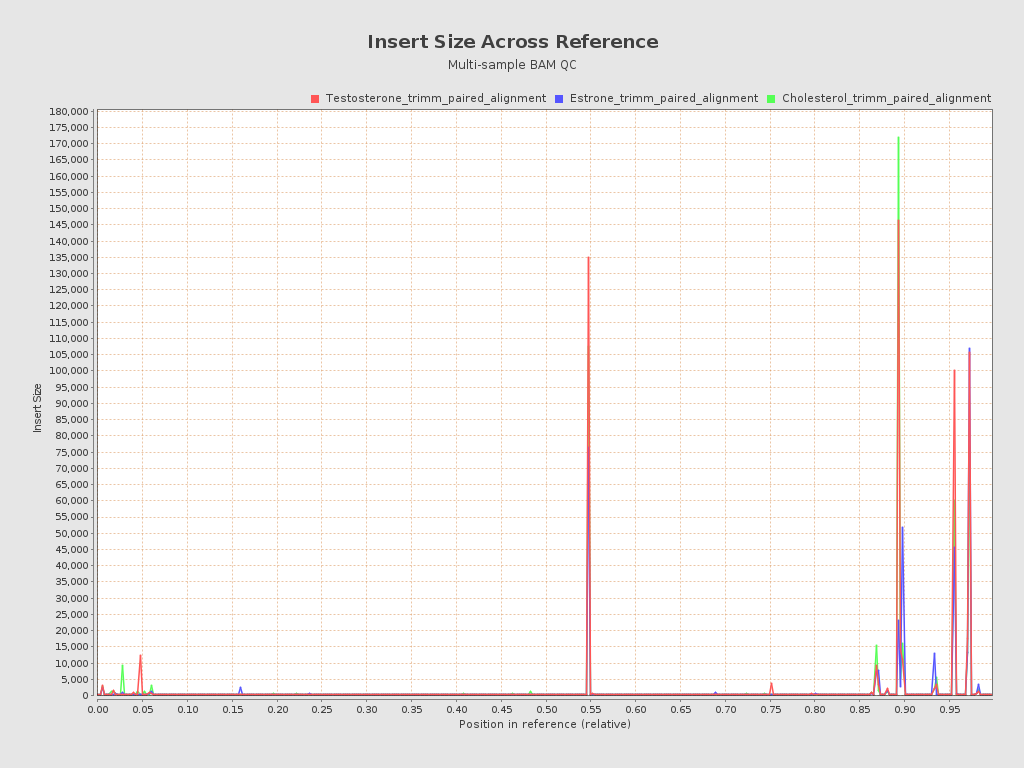
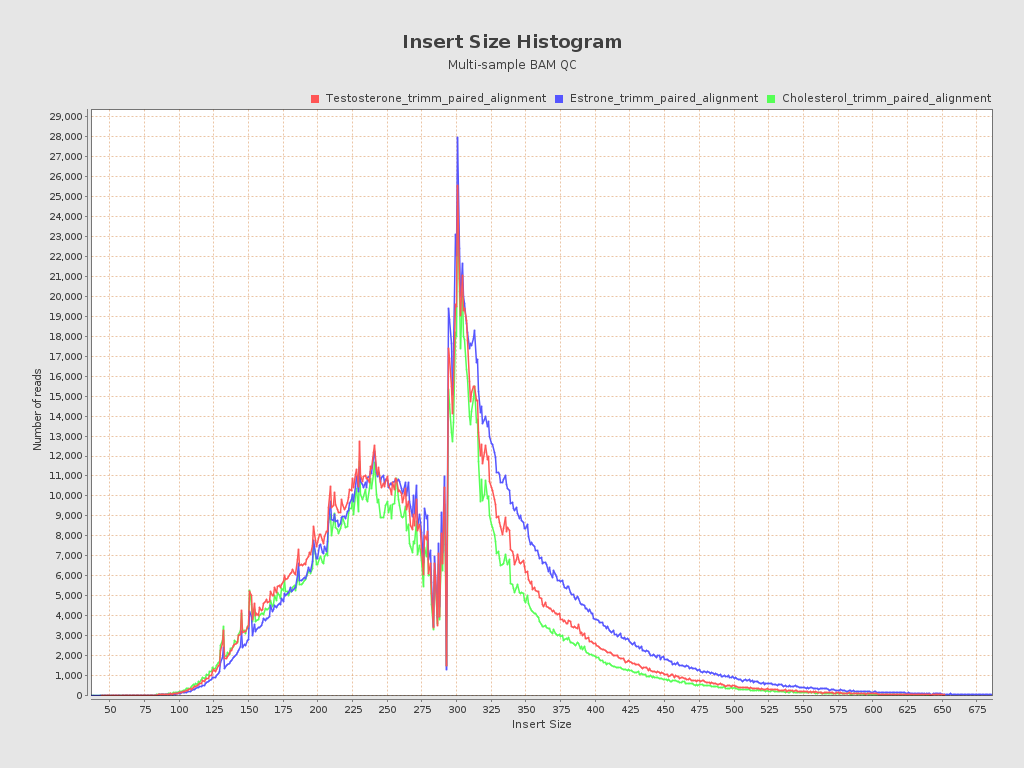
Input data and parameters
Samples
| Testosterone_trimm_paired_alignment | /kb/module/work/tmp/download_f2780a86-43b6-4a18-8032-0ace30e37b6c/accepted_hits_stats |
| Estrone_trimm_paired_alignment | /kb/module/work/tmp/download_ff1ebadb-389a-41b0-ae54-5b4c3d0ad25c/accepted_hits_stats |
| Cholesterol_trimm_paired_alignment | /kb/module/work/tmp/download_4c9202b6-b23b-4276-97d3-2528af1950c0/accepted_hits_stats |
Summary
Globals
| Number of samples | 3 |
| Total number of mapped reads | 52,259,729 |
| Mean samples coverage | 11,512.29 |
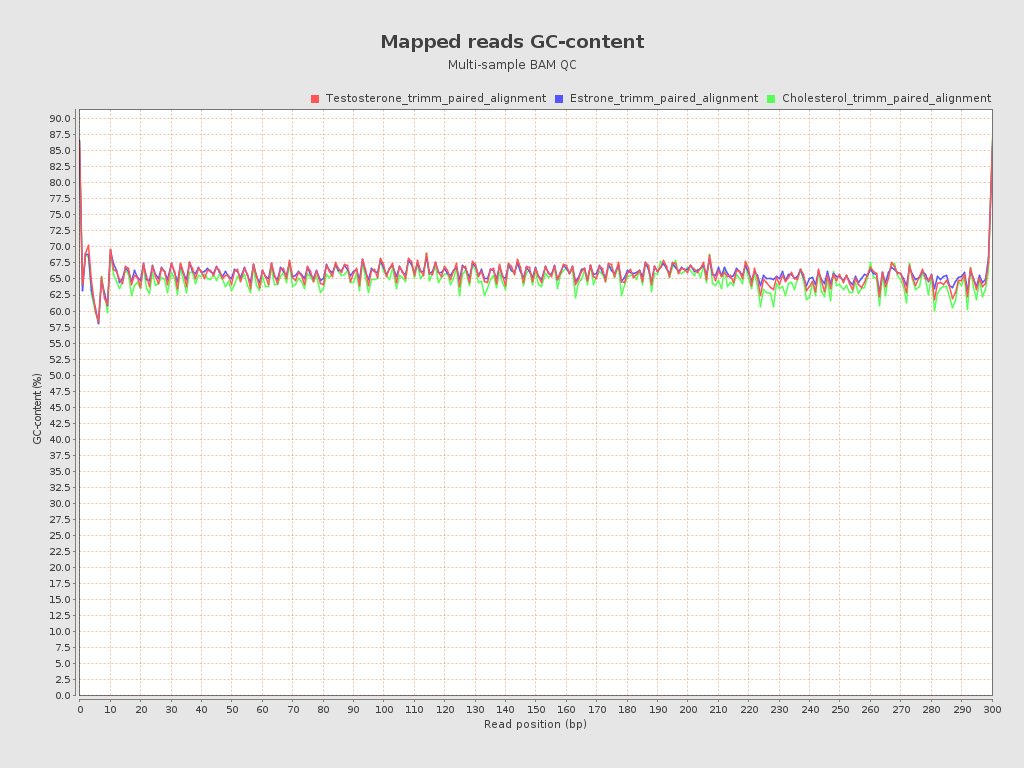
| Mean samples GC-content | 65.75 |
| Mean samples mapping quality | 59.12 |
| Mean samples insert size | 286 |
Sample statistics
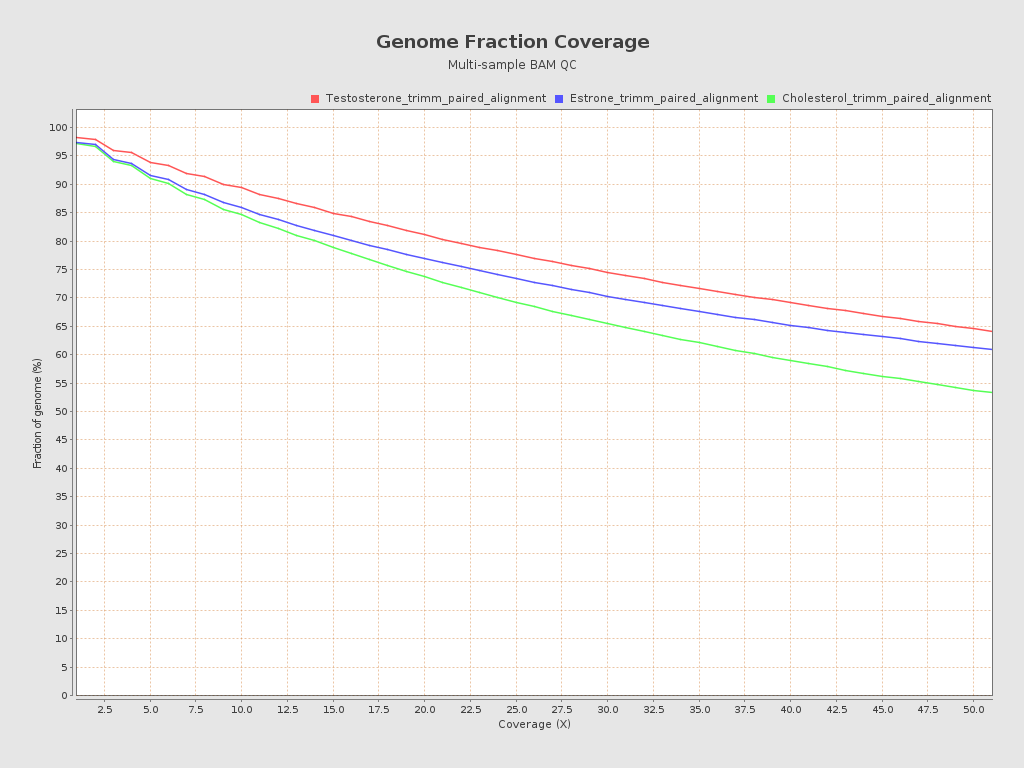
| Sample name | Coverage mean | Coverage std | GC percentage | Mapping quality mean | Insert size median |
| Testosterone_trimm_paired_alignment | 9188.5112 | 16388.8469 | 65.94 | 59.1101 | 281.0 |
| Estrone_trimm_paired_alignment | 6751.9118 | 11361.7389 | 66.17 | 59.1303 | 300.0 |
| Cholesterol_trimm_paired_alignment | 18596.4496 | 26718.9145 | 65.13 | 59.1229 | 277.0 |

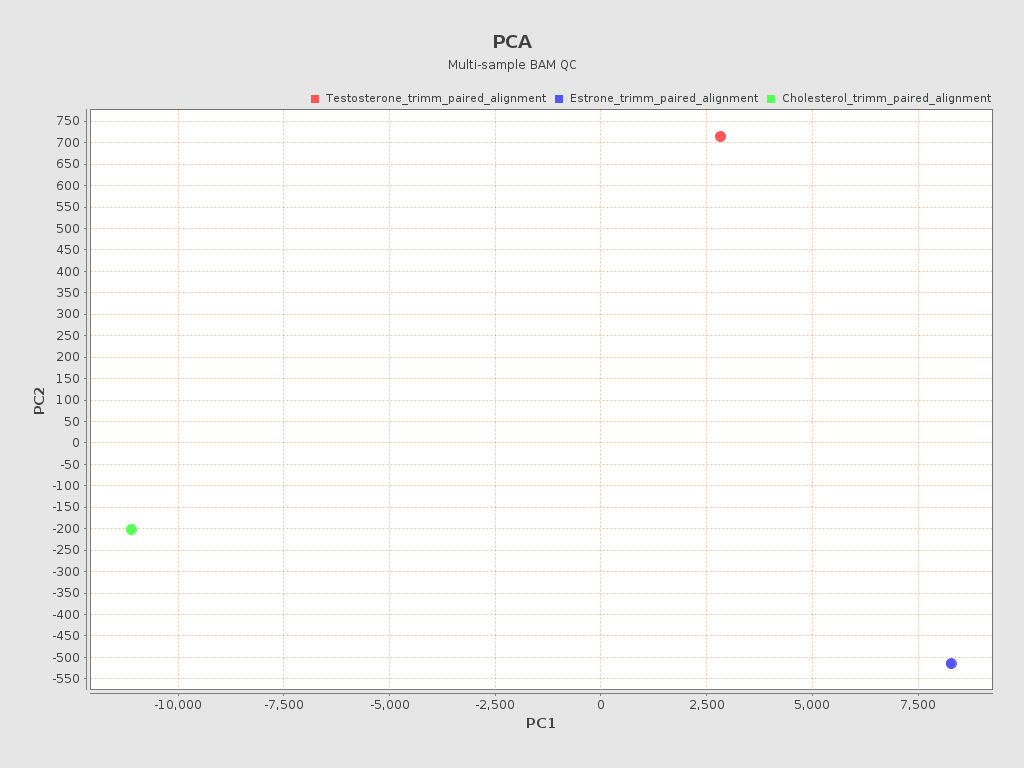
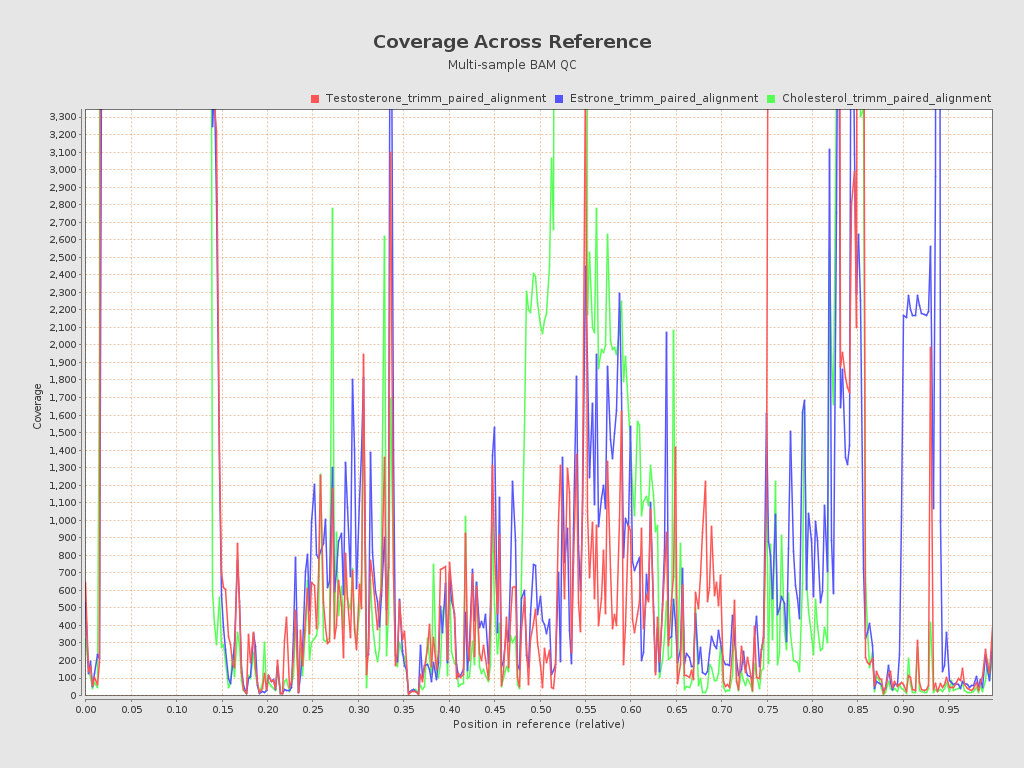
.png)