
ID: growdb:01
Please cite this project and all data from these samples as:
Borton, et al. (2022) GROWdb US River Systems - Samples. [Data set]. DOE Systems Biology Knowledgebase. doi:10.25982/109073.30/1895615.
Genome Resolved Open Watersheds (GROW) is a community effort to increase genomic representation of the world river microbiome with an emphasis on creating an ecosystem-specific publicly available genome database (GROWdb) that will be a resource for microbiologists, geochemists, hydrologists, and modelers and enable new fundamental understandings of watershed microbiology.
DATA AVAILABILITY - requires a KBase login to access:
These data were acquired using a large scale sequencing proposal generated by multiple PIs using samples collected by multiple labs. To protect early career researchers, sample collectors, and data generators, we are asking data users to review and commit to the authorship agreement here: authorship documents
It is our hope that this will 1) protect early career researchers pursuing stories with this dataset 2) maintain attribution for data collectors and generators 3) promote collaboration among research scientists in river corridor microbiomes.
GROWdb contains data from various research campaigns, please acknowledge the following data generators, as appropriate:
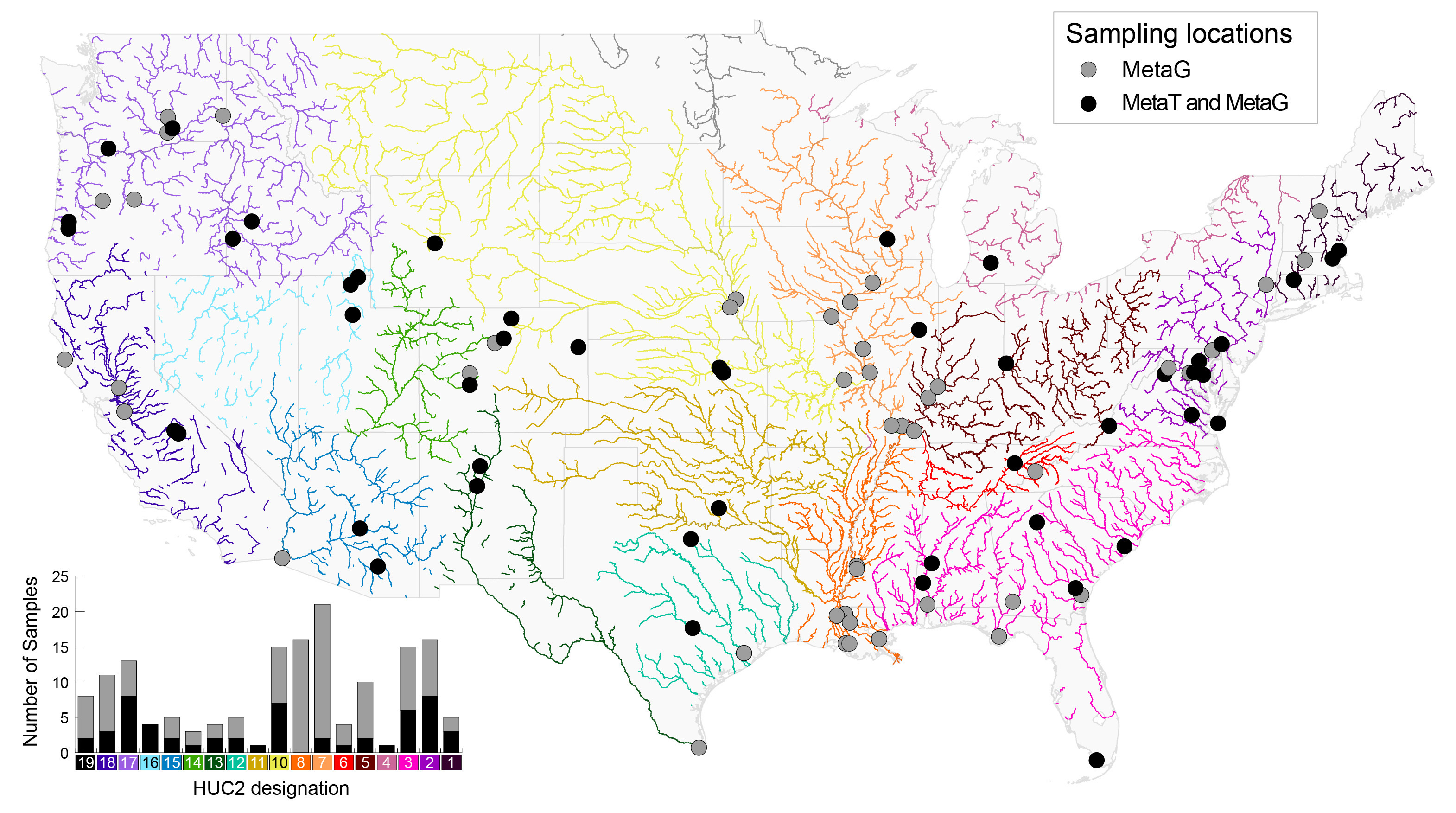
Total Samples loaded onto this Narrative: 178
Note: Not all GROW samples may be loaded into KBase
| Narrative Content | Info | Narrative Link | |
|---|---|---|---|
| GROW MAGs | Dereplicated MAGs 2093 | https://narrative.kbase.us/narrative/106867 | |
| GROW Metabolic Models | Models derived based on MAGs | https://narrative.kbase.us/narrative/107864 |