Nicholas C. Dove, Neslihan Taş, Stephen C. Hart \ [email protected] or [email protected]
Background: Increasing wildfire severity, which is common throughout the western United States, can have deleterious effects on plant regeneration and large impacts on carbon (C) and nitrogen (N) cycling rates. Soil microbes are pivotal in facilitating these elemental cycles, so understanding the impact of increasing fire severity on soil microbial communities is critical.
Methods: We assessed the long-term impact of high-severity fires on the soil microbiome. In the original paper Dove et al. (2022) we we employed high-throughput sequencing of the 16S rRNA gene and the ITS region as well as shotgun metagenomics and reconstruction of microbial genomes (metagenome-assembled genomes--MAGs). We paired these sequence data with previously published soil and vegetation data collected from the Central Sierra Nevada Fire Chronosequence Dove et al. (2020) to elucidate long-term changes in soil microbial structure and function following high-severity wildfires.
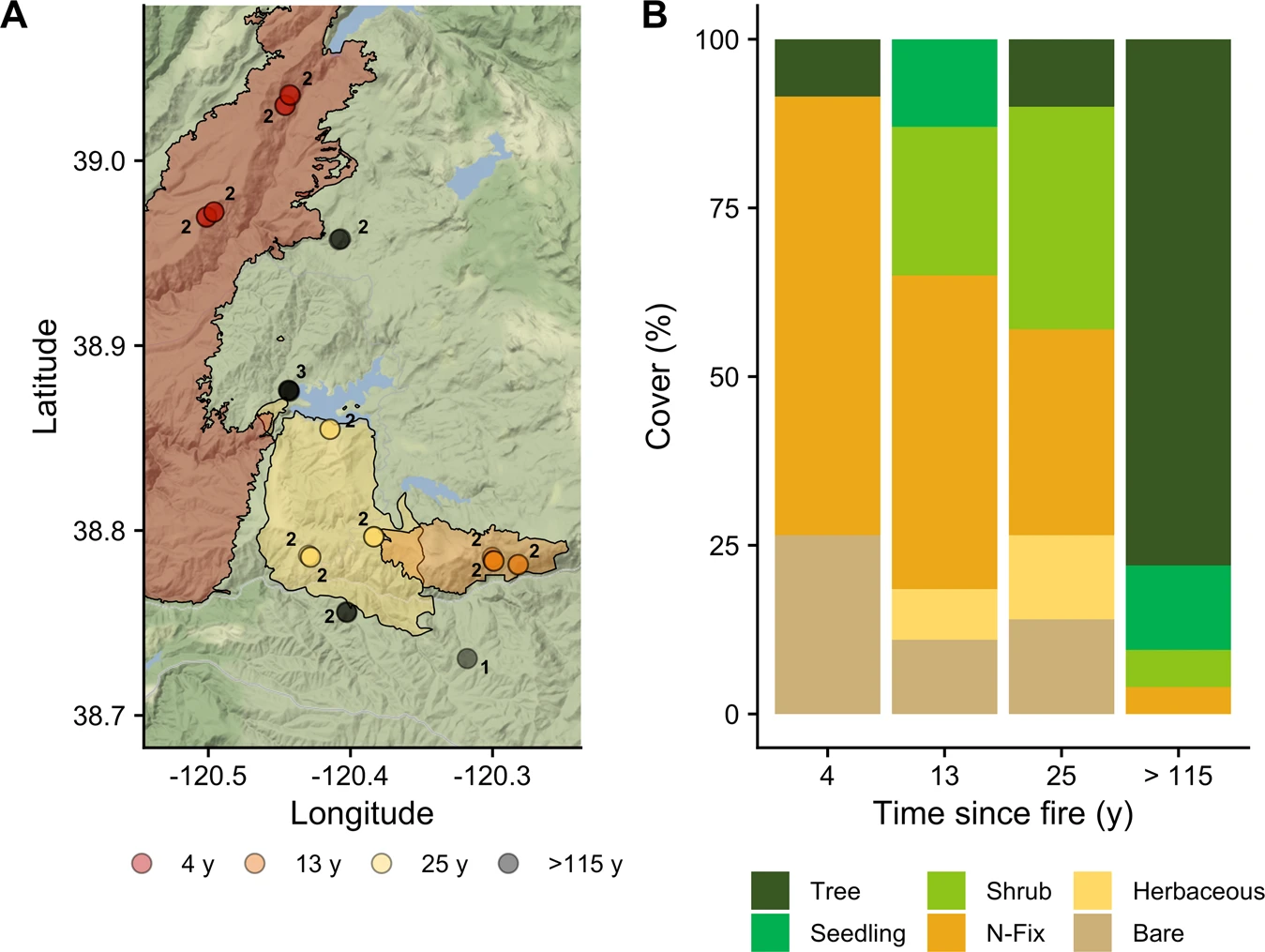
In this narrative we focus on the annotation of MAGs. These MAGs we extracted from 16 metagenomes sequenced from the Central Sierra Nevada Fire Chronosequence (Figure below). Briefly, raw reads were trimmed and quality filtered using Trimmomatic (v 0.36), Samples were co-assembled using MEGAHIT with a minimum contig length of 1000 bp. Then, each individual sample was mapped back to the MEGAHIT contigs with bbmap, and we extracted unmapped reads. Next, unmapped reads were concatenated and re-assembled using SPAdes in the “meta” setting. The newly assembled contig folds were merged with the MEGAHIT contigs. Genome fragments that were larger than 1 kb were clustered into MAGs using MaxBin (v 2.2.5) and MetaBAT2 (v 2.12.1), and MAGs were dereplicated using DASTool (v 1.1.10). Potential mis-binning was identified with CheckM, and bins were further refined to remove contamination following Xue et al. (2020).
Bins were then imported into KBase where they were taxonomically classified with GTDB and functionally annotated with DRAM and RASTtk.
Results: We find that high-severity wildfires result in a multi-decadal (>25 y) recovery of the soil microbiome mediated by concomitant differences in aboveground vegetation, soil chemistry, and microbial assembly processes. Our results depict a distinct taxonomic and functional successional pattern of increasing selection in post-fire soil microbial communities. Changes in microbiome composition corresponded with changes in microbial functional potential, specifically altered C metabolism and enhanced N cycling potential, which related to rates of potential decomposition and inorganic N availability, respectively. Based on MAGs, we show that bacterial genomes enriched in our earliest site (4 y since fire) harbor distinct traits such as a robust stress response and a high potential to degrade pyrogenic, polyaromatic C that allow them to thrive in post-fire environments. Taken together, these results provide a biological basis for previously reported process rate measurements and explain the temporal dynamics of post-fire biogeochemistry, which ultimately constrains ecosystem recovery.

| Created Object Name | Type | Description |
|---|---|---|
| fire_reads_biosamples | SampleSet |

| Created Object Name | Type | Description |
|---|---|---|
| fire_reads_biosamples_clean | SampleSet |

| Created Object Name | Type | Description |
|---|---|---|
| Fire | AssemblySet | Imported Assembly Set |
| CLV.031__Fire_bins | Assembly | Imported Assembly Object |
| CLV.081__Fire_bins | Assembly | Imported Assembly Object |
| CLV.082__Fire_bins | Assembly | Imported Assembly Object |
| CLV.099__Fire_bins | Assembly | Imported Assembly Object |
| CLV.115__Fire_bins | Assembly | Imported Assembly Object |
| CLV.104__Fire_bins | Assembly | Imported Assembly Object |
| CLV.153__Fire_bins | Assembly | Imported Assembly Object |
| CLV.169__Fire_bins | Assembly | Imported Assembly Object |
| CLV.173__Fire_bins | Assembly | Imported Assembly Object |
| CLV.161__Fire_bins | Assembly | Imported Assembly Object |
| CLV.236__Fire_bins | Assembly | Imported Assembly Object |
| CLV.266__Fire_bins | Assembly | Imported Assembly Object |
| CLV.310__Fire_bins | Assembly | Imported Assembly Object |
| CLV.312__Fire_bins | Assembly | Imported Assembly Object |
| CLV.316__Fire_bins | Assembly | Imported Assembly Object |
| CLV.332__Fire_bins | Assembly | Imported Assembly Object |
| CLV.325__Fire_bins | Assembly | Imported Assembly Object |
| CLV.378__Fire_bins | Assembly | Imported Assembly Object |
| CNT.009__Fire_bins | Assembly | Imported Assembly Object |
| CNT.016__Fire_bins | Assembly | Imported Assembly Object |
| CNT.079__Fire_bins | Assembly | Imported Assembly Object |
| CNT.089__Fire_bins | Assembly | Imported Assembly Object |
| CNT.025__Fire_bins | Assembly | Imported Assembly Object |
| CNT.091__Fire_bins | Assembly | Imported Assembly Object |
| CNT.029__Fire_bins | Assembly | Imported Assembly Object |
| CNT.036__Fire_bins | Assembly | Imported Assembly Object |
| CNT.056__Fire_bins | Assembly | Imported Assembly Object |
| CNT.097__Fire_bins | Assembly | Imported Assembly Object |
| CNT.105__Fire_bins | Assembly | Imported Assembly Object |
| CNT.110__Fire_bins | Assembly | Imported Assembly Object |
| CNT.121__Fire_bins | Assembly | Imported Assembly Object |
| CNT.124__Fire_bins | Assembly | Imported Assembly Object |
| CNT.126__Fire_bins | Assembly | Imported Assembly Object |
| CNT.132__Fire_bins | Assembly | Imported Assembly Object |
| CNT.134__Fire_bins | Assembly | Imported Assembly Object |
| CNT.137__Fire_bins | Assembly | Imported Assembly Object |
| CNT.150__Fire_bins | Assembly | Imported Assembly Object |
| CNT.161__Fire_bins | Assembly | Imported Assembly Object |
| CNT.186__Fire_bins | Assembly | Imported Assembly Object |
| CNT.188__Fire_bins | Assembly | Imported Assembly Object |
| CNT.191__Fire_bins | Assembly | Imported Assembly Object |
| CNT.275__Fire_bins | Assembly | Imported Assembly Object |
| CNT.192__Fire_bins | Assembly | Imported Assembly Object |
| CNT.193__Fire_bins | Assembly | Imported Assembly Object |
| CNT.195__Fire_bins | Assembly | Imported Assembly Object |
| CNT.279__Fire_bins | Assembly | Imported Assembly Object |
| CNT.212__Fire_bins | Assembly | Imported Assembly Object |
| CNT.271__Fire_bins | Assembly | Imported Assembly Object |
| CNT.259__Fire_bins | Assembly | Imported Assembly Object |
| CNT.284__Fire_bins | Assembly | Imported Assembly Object |
| CNT.289__Fire_bins | Assembly | Imported Assembly Object |
| CNT.293__Fire_bins | Assembly | Imported Assembly Object |
| CNT.308__Fire_bins | Assembly | Imported Assembly Object |
| CNT.312__Fire_bins | Assembly | Imported Assembly Object |
| CNT.331__Fire_bins | Assembly | Imported Assembly Object |
| CNT.345__Fire_bins | Assembly | Imported Assembly Object |
| CNT.381__Fire_bins | Assembly | Imported Assembly Object |
| CNT.405__Fire_bins | Assembly | Imported Assembly Object |
| CNT.333__Fire_bins | Assembly | Imported Assembly Object |
| CNT.435__Fire_bins | Assembly | Imported Assembly Object |
| CNT.531__Fire_bins | Assembly | Imported Assembly Object |
| CNT.536__Fire_bins | Assembly | Imported Assembly Object |
| CNT.447__Fire_bins | Assembly | Imported Assembly Object |
| FDR.001__Fire_bins | Assembly | Imported Assembly Object |
| CNT.474__Fire_bins | Assembly | Imported Assembly Object |
| CNT.470__Fire_bins | Assembly | Imported Assembly Object |
| FDR.010__Fire_bins | Assembly | Imported Assembly Object |
| CNT.528__Fire_bins | Assembly | Imported Assembly Object |
| FDR.018__Fire_bins | Assembly | Imported Assembly Object |
| FDR.048__Fire_bins | Assembly | Imported Assembly Object |
| FDR.073__Fire_bins | Assembly | Imported Assembly Object |
| FDR.075__Fire_bins | Assembly | Imported Assembly Object |
| FDR.101__Fire_bins | Assembly | Imported Assembly Object |
| FDR.112__Fire_bins | Assembly | Imported Assembly Object |
| FDR.171__Fire_bins | Assembly | Imported Assembly Object |
| FDR.178__Fire_bins | Assembly | Imported Assembly Object |
| FDR.187__Fire_bins | Assembly | Imported Assembly Object |
| FDR.211__Fire_bins | Assembly | Imported Assembly Object |
| FDR.226__Fire_bins | Assembly | Imported Assembly Object |
| FDR.307__Fire_bins | Assembly | Imported Assembly Object |
| FDR.312__Fire_bins | Assembly | Imported Assembly Object |
| FDR.338__Fire_bins | Assembly | Imported Assembly Object |
| FDR.348__Fire_bins | Assembly | Imported Assembly Object |
| FDR.480__Fire_bins | Assembly | Imported Assembly Object |
| FDR.359__Fire_bins | Assembly | Imported Assembly Object |
| FDR.407__Fire_bins | Assembly | Imported Assembly Object |
| FDR.416__Fire_bins | Assembly | Imported Assembly Object |
| FDR.436__Fire_bins | Assembly | Imported Assembly Object |
| FDR.444__Fire_bins | Assembly | Imported Assembly Object |
| FDR.445__Fire_bins | Assembly | Imported Assembly Object |
| FDR.489__Fire_bins | Assembly | Imported Assembly Object |
| FDR.500__Fire_bins | Assembly | Imported Assembly Object |
| FDR.501__Fire_bins | Assembly | Imported Assembly Object |
| FDR.502__Fire_bins | Assembly | Imported Assembly Object |
| FDR.512__Fire_bins | Assembly | Imported Assembly Object |
| FDR.521__Fire_bins | Assembly | Imported Assembly Object |
| FDR.522__Fire_bins | Assembly | Imported Assembly Object |
| FDR.549__Fire_bins | Assembly | Imported Assembly Object |
| FDR.554__Fire_bins | Assembly | Imported Assembly Object |
| FDR.556__Fire_bins | Assembly | Imported Assembly Object |
| FDR.560__Fire_bins | Assembly | Imported Assembly Object |
| FDR.595__Fire_bins | Assembly | Imported Assembly Object |
| FDR.566__Fire_bins | Assembly | Imported Assembly Object |
| FDR.606__Fire_bins | Assembly | Imported Assembly Object |
| KNG.005__Fire_bins | Assembly | Imported Assembly Object |
| FDR.625__Fire_bins | Assembly | Imported Assembly Object |
| KNG.006__Fire_bins | Assembly | Imported Assembly Object |
| FDR.628__Fire_bins | Assembly | Imported Assembly Object |
| FDR.620__Fire_bins | Assembly | Imported Assembly Object |
| KNG.009__Fire_bins | Assembly | Imported Assembly Object |
| FDR.614__Fire_bins | Assembly | Imported Assembly Object |
| KNG.011__Fire_bins | Assembly | Imported Assembly Object |
| KNG.003__Fire_bins | Assembly | Imported Assembly Object |
| KNG.012__Fire_bins | Assembly | Imported Assembly Object |
| KNG.013__Fire_bins | Assembly | Imported Assembly Object |
| KNG.014__Fire_bins | Assembly | Imported Assembly Object |
| KNG.015__Fire_bins | Assembly | Imported Assembly Object |
| KNG.020__Fire_bins | Assembly | Imported Assembly Object |
| KNG.027__Fire_bins | Assembly | Imported Assembly Object |
| KNG.033__Fire_bins | Assembly | Imported Assembly Object |
| KNG.044__Fire_bins | Assembly | Imported Assembly Object |
| KNG.046__Fire_bins | Assembly | Imported Assembly Object |
| KNG.051__Fire_bins | Assembly | Imported Assembly Object |
| KNG.058__Fire_bins | Assembly | Imported Assembly Object |
| KNG.061__Fire_bins | Assembly | Imported Assembly Object |
| KNG.080__Fire_bins | Assembly | Imported Assembly Object |
| KNG.083__Fire_bins | Assembly | Imported Assembly Object |
| KNG.085__Fire_bins | Assembly | Imported Assembly Object |
| KNG.135__Fire_bins | Assembly | Imported Assembly Object |
| KNG.140__Fire_bins | Assembly | Imported Assembly Object |
| KNG.108__Fire_bins | Assembly | Imported Assembly Object |
| KNG.118__Fire_bins | Assembly | Imported Assembly Object |
| KNG.120__Fire_bins | Assembly | Imported Assembly Object |
| KNG.146__Fire_bins | Assembly | Imported Assembly Object |
| KNG.159__Fire_bins | Assembly | Imported Assembly Object |
| KNG.165__Fire_bins | Assembly | Imported Assembly Object |
| KNG.178__Fire_bins | Assembly | Imported Assembly Object |
| KNG.177__Fire_bins | Assembly | Imported Assembly Object |
| KNG.186__Fire_bins | Assembly | Imported Assembly Object |
| KNG.190__Fire_bins | Assembly | Imported Assembly Object |
| KNG.192__Fire_bins | Assembly | Imported Assembly Object |
| KNG.195__Fire_bins | Assembly | Imported Assembly Object |
| KNG.210__Fire_bins | Assembly | Imported Assembly Object |
| KNG.216__Fire_bins | Assembly | Imported Assembly Object |
| KNG.219__Fire_bins | Assembly | Imported Assembly Object |
| KNG.225__Fire_bins | Assembly | Imported Assembly Object |
| KNG.262__Fire_bins | Assembly | Imported Assembly Object |
| KNG.226__Fire_bins | Assembly | Imported Assembly Object |
| KNG.281__Fire_bins | Assembly | Imported Assembly Object |
| KNG.284__Fire_bins | Assembly | Imported Assembly Object |
| KNG.244__Fire_bins | Assembly | Imported Assembly Object |
| KNG.247__Fire_bins | Assembly | Imported Assembly Object |
| KNG.299__Fire_bins | Assembly | Imported Assembly Object |
| KNG.252__Fire_bins | Assembly | Imported Assembly Object |
| KNG.259__Fire_bins | Assembly | Imported Assembly Object |
| KNG.302__Fire_bins | Assembly | Imported Assembly Object |
| KNG.305__Fire_bins | Assembly | Imported Assembly Object |
| KNG.307__Fire_bins | Assembly | Imported Assembly Object |
| KNG.316__Fire_bins | Assembly | Imported Assembly Object |
| KNG.322__Fire_bins | Assembly | Imported Assembly Object |
| KNG.324__Fire_bins | Assembly | Imported Assembly Object |
| KNG.331__Fire_bins | Assembly | Imported Assembly Object |
| KNG.339__Fire_bins | Assembly | Imported Assembly Object |
| KNG.348__Fire_bins | Assembly | Imported Assembly Object |
| KNG.392__Fire_bins | Assembly | Imported Assembly Object |
| KNG.402__Fire_bins | Assembly | Imported Assembly Object |
| KNG.408__Fire_bins | Assembly | Imported Assembly Object |
| KNG.421__Fire_bins | Assembly | Imported Assembly Object |
| KNG.448__Fire_bins | Assembly | Imported Assembly Object |
| KNG.571__Fire_bins | Assembly | Imported Assembly Object |
| KNG.578__Fire_bins | Assembly | Imported Assembly Object |
| KNG.514__Fire_bins | Assembly | Imported Assembly Object |
| KNG.534__Fire_bins | Assembly | Imported Assembly Object |
| KNG.610__Fire_bins | Assembly | Imported Assembly Object |
| KNG.540__Fire_bins | Assembly | Imported Assembly Object |
| KNG.559__Fire_bins | Assembly | Imported Assembly Object |
| KNG.566__Fire_bins | Assembly | Imported Assembly Object |
| KNG.645__Fire_bins | Assembly | Imported Assembly Object |
| KNG.653__Fire_bins | Assembly | Imported Assembly Object |
| KNG.670__Fire_bins | Assembly | Imported Assembly Object |
| KNG.688__Fire_bins | Assembly | Imported Assembly Object |
| KNG.699__Fire_bins | Assembly | Imported Assembly Object |
| KNG.710__Fire_bins | Assembly | Imported Assembly Object |
| KNG.716__Fire_bins | Assembly | Imported Assembly Object |
| KNG.728__Fire_bins | Assembly | Imported Assembly Object |
| KNG.732__Fire_bins | Assembly | Imported Assembly Object |
| KNG.740__Fire_bins | Assembly | Imported Assembly Object |
| KNG.747__Fire_bins | Assembly | Imported Assembly Object |
| KNG.761__Fire_bins | Assembly | Imported Assembly Object |
| KNG.766__Fire_bins | Assembly | Imported Assembly Object |
| KNG.774__Fire_bins | Assembly | Imported Assembly Object |
| KNG.813__Fire_bins | Assembly | Imported Assembly Object |
| KNG.793__Fire_bins | Assembly | Imported Assembly Object |
| KNG.794__Fire_bins | Assembly | Imported Assembly Object |
| KNG.799__Fire_bins | Assembly | Imported Assembly Object |
| KNG.800__Fire_bins | Assembly | Imported Assembly Object |
| KNG.802__Fire_bins | Assembly | Imported Assembly Object |
| KNG.807__Fire_bins | Assembly | Imported Assembly Object |
| KNG.820__Fire_bins | Assembly | Imported Assembly Object |
| KNG.826__Fire_bins | Assembly | Imported Assembly Object |
| KNG.828__Fire_bins | Assembly | Imported Assembly Object |
| KNG.829__Fire_bins | Assembly | Imported Assembly Object |
| KNG.830__Fire_bins | Assembly | Imported Assembly Object |
| KNG.832__Fire_bins | Assembly | Imported Assembly Object |
| KNG.833__Fire_bins | Assembly | Imported Assembly Object |
| Created Object Name | Type | Description |
|---|---|---|
| CLV.031__Fire_bins_genome | Genome | Annotated Genome |
| CLV.081__Fire_bins_genome | Genome | Annotated Genome |
| CLV.082__Fire_bins_genome | Genome | Annotated Genome |
| CLV.099__Fire_bins_genome | Genome | Annotated Genome |
| CLV.104__Fire_bins_genome | Genome | Annotated Genome |
| CLV.115__Fire_bins_genome | Genome | Annotated Genome |
| CLV.153__Fire_bins_genome | Genome | Annotated Genome |
| CLV.161__Fire_bins_genome | Genome | Annotated Genome |
| CLV.169__Fire_bins_genome | Genome | Annotated Genome |
| CLV.173__Fire_bins_genome | Genome | Annotated Genome |
| CLV.236__Fire_bins_genome | Genome | Annotated Genome |
| CLV.266__Fire_bins_genome | Genome | Annotated Genome |
| CLV.310__Fire_bins_genome | Genome | Annotated Genome |
| CLV.312__Fire_bins_genome | Genome | Annotated Genome |
| CLV.316__Fire_bins_genome | Genome | Annotated Genome |
| CLV.325__Fire_bins_genome | Genome | Annotated Genome |
| CLV.332__Fire_bins_genome | Genome | Annotated Genome |
| CLV.378__Fire_bins_genome | Genome | Annotated Genome |
| CNT.009__Fire_bins_genome | Genome | Annotated Genome |
| CNT.016__Fire_bins_genome | Genome | Annotated Genome |
| CNT.025__Fire_bins_genome | Genome | Annotated Genome |
| CNT.029__Fire_bins_genome | Genome | Annotated Genome |
| CNT.036__Fire_bins_genome | Genome | Annotated Genome |
| CNT.056__Fire_bins_genome | Genome | Annotated Genome |
| CNT.079__Fire_bins_genome | Genome | Annotated Genome |
| CNT.089__Fire_bins_genome | Genome | Annotated Genome |
| CNT.091__Fire_bins_genome | Genome | Annotated Genome |
| CNT.097__Fire_bins_genome | Genome | Annotated Genome |
| CNT.105__Fire_bins_genome | Genome | Annotated Genome |
| CNT.110__Fire_bins_genome | Genome | Annotated Genome |
| CNT.121__Fire_bins_genome | Genome | Annotated Genome |
| CNT.124__Fire_bins_genome | Genome | Annotated Genome |
| CNT.126__Fire_bins_genome | Genome | Annotated Genome |
| CNT.132__Fire_bins_genome | Genome | Annotated Genome |
| CNT.134__Fire_bins_genome | Genome | Annotated Genome |
| CNT.137__Fire_bins_genome | Genome | Annotated Genome |
| CNT.150__Fire_bins_genome | Genome | Annotated Genome |
| CNT.161__Fire_bins_genome | Genome | Annotated Genome |
| CNT.186__Fire_bins_genome | Genome | Annotated Genome |
| CNT.188__Fire_bins_genome | Genome | Annotated Genome |
| CNT.191__Fire_bins_genome | Genome | Annotated Genome |
| CNT.192__Fire_bins_genome | Genome | Annotated Genome |
| CNT.193__Fire_bins_genome | Genome | Annotated Genome |
| CNT.195__Fire_bins_genome | Genome | Annotated Genome |
| CNT.212__Fire_bins_genome | Genome | Annotated Genome |
| CNT.259__Fire_bins_genome | Genome | Annotated Genome |
| CNT.271__Fire_bins_genome | Genome | Annotated Genome |
| CNT.275__Fire_bins_genome | Genome | Annotated Genome |
| CNT.279__Fire_bins_genome | Genome | Annotated Genome |
| CNT.284__Fire_bins_genome | Genome | Annotated Genome |
| CNT.289__Fire_bins_genome | Genome | Annotated Genome |
| CNT.293__Fire_bins_genome | Genome | Annotated Genome |
| CNT.308__Fire_bins_genome | Genome | Annotated Genome |
| CNT.312__Fire_bins_genome | Genome | Annotated Genome |
| CNT.331__Fire_bins_genome | Genome | Annotated Genome |
| CNT.333__Fire_bins_genome | Genome | Annotated Genome |
| CNT.345__Fire_bins_genome | Genome | Annotated Genome |
| CNT.381__Fire_bins_genome | Genome | Annotated Genome |
| CNT.405__Fire_bins_genome | Genome | Annotated Genome |
| CNT.435__Fire_bins_genome | Genome | Annotated Genome |
| CNT.447__Fire_bins_genome | Genome | Annotated Genome |
| CNT.470__Fire_bins_genome | Genome | Annotated Genome |
| CNT.474__Fire_bins_genome | Genome | Annotated Genome |
| CNT.528__Fire_bins_genome | Genome | Annotated Genome |
| CNT.531__Fire_bins_genome | Genome | Annotated Genome |
| CNT.536__Fire_bins_genome | Genome | Annotated Genome |
| FDR.001__Fire_bins_genome | Genome | Annotated Genome |
| FDR.010__Fire_bins_genome | Genome | Annotated Genome |
| FDR.018__Fire_bins_genome | Genome | Annotated Genome |
| FDR.048__Fire_bins_genome | Genome | Annotated Genome |
| FDR.073__Fire_bins_genome | Genome | Annotated Genome |
| FDR.075__Fire_bins_genome | Genome | Annotated Genome |
| FDR.101__Fire_bins_genome | Genome | Annotated Genome |
| FDR.112__Fire_bins_genome | Genome | Annotated Genome |
| FDR.171__Fire_bins_genome | Genome | Annotated Genome |
| FDR.178__Fire_bins_genome | Genome | Annotated Genome |
| FDR.187__Fire_bins_genome | Genome | Annotated Genome |
| FDR.211__Fire_bins_genome | Genome | Annotated Genome |
| FDR.226__Fire_bins_genome | Genome | Annotated Genome |
| FDR.307__Fire_bins_genome | Genome | Annotated Genome |
| FDR.312__Fire_bins_genome | Genome | Annotated Genome |
| FDR.338__Fire_bins_genome | Genome | Annotated Genome |
| FDR.348__Fire_bins_genome | Genome | Annotated Genome |
| FDR.359__Fire_bins_genome | Genome | Annotated Genome |
| FDR.407__Fire_bins_genome | Genome | Annotated Genome |
| FDR.416__Fire_bins_genome | Genome | Annotated Genome |
| FDR.436__Fire_bins_genome | Genome | Annotated Genome |
| FDR.444__Fire_bins_genome | Genome | Annotated Genome |
| FDR.445__Fire_bins_genome | Genome | Annotated Genome |
| FDR.480__Fire_bins_genome | Genome | Annotated Genome |
| FDR.489__Fire_bins_genome | Genome | Annotated Genome |
| FDR.500__Fire_bins_genome | Genome | Annotated Genome |
| FDR.501__Fire_bins_genome | Genome | Annotated Genome |
| FDR.502__Fire_bins_genome | Genome | Annotated Genome |
| FDR.512__Fire_bins_genome | Genome | Annotated Genome |
| FDR.521__Fire_bins_genome | Genome | Annotated Genome |
| FDR.522__Fire_bins_genome | Genome | Annotated Genome |
| FDR.549__Fire_bins_genome | Genome | Annotated Genome |
| FDR.554__Fire_bins_genome | Genome | Annotated Genome |
| FDR.556__Fire_bins_genome | Genome | Annotated Genome |
| FDR.560__Fire_bins_genome | Genome | Annotated Genome |
| FDR.566__Fire_bins_genome | Genome | Annotated Genome |
| FDR.595__Fire_bins_genome | Genome | Annotated Genome |
| FDR.606__Fire_bins_genome | Genome | Annotated Genome |
| FDR.614__Fire_bins_genome | Genome | Annotated Genome |
| FDR.620__Fire_bins_genome | Genome | Annotated Genome |
| FDR.625__Fire_bins_genome | Genome | Annotated Genome |
| FDR.628__Fire_bins_genome | Genome | Annotated Genome |
| KNG.003__Fire_bins_genome | Genome | Annotated Genome |
| KNG.005__Fire_bins_genome | Genome | Annotated Genome |
| KNG.006__Fire_bins_genome | Genome | Annotated Genome |
| KNG.009__Fire_bins_genome | Genome | Annotated Genome |
| KNG.011__Fire_bins_genome | Genome | Annotated Genome |
| KNG.012__Fire_bins_genome | Genome | Annotated Genome |
| KNG.013__Fire_bins_genome | Genome | Annotated Genome |
| KNG.014__Fire_bins_genome | Genome | Annotated Genome |
| KNG.015__Fire_bins_genome | Genome | Annotated Genome |
| KNG.020__Fire_bins_genome | Genome | Annotated Genome |
| KNG.027__Fire_bins_genome | Genome | Annotated Genome |
| KNG.033__Fire_bins_genome | Genome | Annotated Genome |
| KNG.044__Fire_bins_genome | Genome | Annotated Genome |
| KNG.046__Fire_bins_genome | Genome | Annotated Genome |
| KNG.051__Fire_bins_genome | Genome | Annotated Genome |
| KNG.058__Fire_bins_genome | Genome | Annotated Genome |
| KNG.061__Fire_bins_genome | Genome | Annotated Genome |
| KNG.080__Fire_bins_genome | Genome | Annotated Genome |
| KNG.083__Fire_bins_genome | Genome | Annotated Genome |
| KNG.085__Fire_bins_genome | Genome | Annotated Genome |
| KNG.108__Fire_bins_genome | Genome | Annotated Genome |
| KNG.118__Fire_bins_genome | Genome | Annotated Genome |
| KNG.120__Fire_bins_genome | Genome | Annotated Genome |
| KNG.135__Fire_bins_genome | Genome | Annotated Genome |
| KNG.140__Fire_bins_genome | Genome | Annotated Genome |
| KNG.146__Fire_bins_genome | Genome | Annotated Genome |
| KNG.159__Fire_bins_genome | Genome | Annotated Genome |
| KNG.165__Fire_bins_genome | Genome | Annotated Genome |
| KNG.177__Fire_bins_genome | Genome | Annotated Genome |
| KNG.178__Fire_bins_genome | Genome | Annotated Genome |
| KNG.186__Fire_bins_genome | Genome | Annotated Genome |
| KNG.190__Fire_bins_genome | Genome | Annotated Genome |
| KNG.192__Fire_bins_genome | Genome | Annotated Genome |
| KNG.195__Fire_bins_genome | Genome | Annotated Genome |
| KNG.210__Fire_bins_genome | Genome | Annotated Genome |
| KNG.216__Fire_bins_genome | Genome | Annotated Genome |
| KNG.219__Fire_bins_genome | Genome | Annotated Genome |
| KNG.225__Fire_bins_genome | Genome | Annotated Genome |
| KNG.226__Fire_bins_genome | Genome | Annotated Genome |
| KNG.244__Fire_bins_genome | Genome | Annotated Genome |
| KNG.247__Fire_bins_genome | Genome | Annotated Genome |
| KNG.252__Fire_bins_genome | Genome | Annotated Genome |
| KNG.259__Fire_bins_genome | Genome | Annotated Genome |
| KNG.262__Fire_bins_genome | Genome | Annotated Genome |
| KNG.281__Fire_bins_genome | Genome | Annotated Genome |
| KNG.284__Fire_bins_genome | Genome | Annotated Genome |
| KNG.299__Fire_bins_genome | Genome | Annotated Genome |
| KNG.302__Fire_bins_genome | Genome | Annotated Genome |
| KNG.305__Fire_bins_genome | Genome | Annotated Genome |
| KNG.307__Fire_bins_genome | Genome | Annotated Genome |
| KNG.316__Fire_bins_genome | Genome | Annotated Genome |
| KNG.322__Fire_bins_genome | Genome | Annotated Genome |
| KNG.324__Fire_bins_genome | Genome | Annotated Genome |
| KNG.331__Fire_bins_genome | Genome | Annotated Genome |
| KNG.339__Fire_bins_genome | Genome | Annotated Genome |
| KNG.348__Fire_bins_genome | Genome | Annotated Genome |
| KNG.392__Fire_bins_genome | Genome | Annotated Genome |
| KNG.402__Fire_bins_genome | Genome | Annotated Genome |
| KNG.408__Fire_bins_genome | Genome | Annotated Genome |
| KNG.421__Fire_bins_genome | Genome | Annotated Genome |
| KNG.448__Fire_bins_genome | Genome | Annotated Genome |
| KNG.514__Fire_bins_genome | Genome | Annotated Genome |
| KNG.534__Fire_bins_genome | Genome | Annotated Genome |
| KNG.540__Fire_bins_genome | Genome | Annotated Genome |
| KNG.559__Fire_bins_genome | Genome | Annotated Genome |
| KNG.566__Fire_bins_genome | Genome | Annotated Genome |
| KNG.571__Fire_bins_genome | Genome | Annotated Genome |
| KNG.578__Fire_bins_genome | Genome | Annotated Genome |
| KNG.610__Fire_bins_genome | Genome | Annotated Genome |
| KNG.645__Fire_bins_genome | Genome | Annotated Genome |
| KNG.653__Fire_bins_genome | Genome | Annotated Genome |
| KNG.670__Fire_bins_genome | Genome | Annotated Genome |
| KNG.688__Fire_bins_genome | Genome | Annotated Genome |
| KNG.699__Fire_bins_genome | Genome | Annotated Genome |
| KNG.710__Fire_bins_genome | Genome | Annotated Genome |
| KNG.716__Fire_bins_genome | Genome | Annotated Genome |
| KNG.728__Fire_bins_genome | Genome | Annotated Genome |
| KNG.732__Fire_bins_genome | Genome | Annotated Genome |
| KNG.740__Fire_bins_genome | Genome | Annotated Genome |
| KNG.747__Fire_bins_genome | Genome | Annotated Genome |
| KNG.761__Fire_bins_genome | Genome | Annotated Genome |
| KNG.766__Fire_bins_genome | Genome | Annotated Genome |
| KNG.774__Fire_bins_genome | Genome | Annotated Genome |
| KNG.793__Fire_bins_genome | Genome | Annotated Genome |
| KNG.794__Fire_bins_genome | Genome | Annotated Genome |
| KNG.799__Fire_bins_genome | Genome | Annotated Genome |
| KNG.800__Fire_bins_genome | Genome | Annotated Genome |
| KNG.802__Fire_bins_genome | Genome | Annotated Genome |
| KNG.807__Fire_bins_genome | Genome | Annotated Genome |
| KNG.813__Fire_bins_genome | Genome | Annotated Genome |
| KNG.820__Fire_bins_genome | Genome | Annotated Genome |
| KNG.826__Fire_bins_genome | Genome | Annotated Genome |
| KNG.828__Fire_bins_genome | Genome | Annotated Genome |
| KNG.829__Fire_bins_genome | Genome | Annotated Genome |
| KNG.830__Fire_bins_genome | Genome | Annotated Genome |
| KNG.832__Fire_bins_genome | Genome | Annotated Genome |
| KNG.833__Fire_bins_genome | Genome | Annotated Genome |
| Fire_Dram | GenomeSet | Fire |

| Fire |
| Created Object Name | Type | Description |
|---|---|---|
| KNG.011__Fire_bins.RAST | Genome | RAST annotation |
| KNG.159__Fire_bins.RAST | Genome | RAST annotation |
| KNG.307__Fire_bins.RAST | Genome | RAST annotation |
| CNT.259__Fire_bins.RAST | Genome | RAST annotation |
| FDR.075__Fire_bins.RAST | Genome | RAST annotation |
| FDR.512__Fire_bins.RAST | Genome | RAST annotation |
| KNG.284__Fire_bins.RAST | Genome | RAST annotation |
| CNT.132__Fire_bins.RAST | Genome | RAST annotation |
| CNT.381__Fire_bins.RAST | Genome | RAST annotation |
| CLV.236__Fire_bins.RAST | Genome | RAST annotation |
| KNG.324__Fire_bins.RAST | Genome | RAST annotation |
| KNG.716__Fire_bins.RAST | Genome | RAST annotation |
| KNG.225__Fire_bins.RAST | Genome | RAST annotation |
| FDR.556__Fire_bins.RAST | Genome | RAST annotation |
| KNG.051__Fire_bins.RAST | Genome | RAST annotation |
| FDR.348__Fire_bins.RAST | Genome | RAST annotation |
| CNT.435__Fire_bins.RAST | Genome | RAST annotation |
| CNT.195__Fire_bins.RAST | Genome | RAST annotation |
| CNT.528__Fire_bins.RAST | Genome | RAST annotation |
| CNT.089__Fire_bins.RAST | Genome | RAST annotation |
| KNG.534__Fire_bins.RAST | Genome | RAST annotation |
| KNG.800__Fire_bins.RAST | Genome | RAST annotation |
| CLV.153__Fire_bins.RAST | Genome | RAST annotation |
| KNG.165__Fire_bins.RAST | Genome | RAST annotation |
| KNG.003__Fire_bins.RAST | Genome | RAST annotation |
| FDR.502__Fire_bins.RAST | Genome | RAST annotation |
| FDR.073__Fire_bins.RAST | Genome | RAST annotation |
| KNG.244__Fire_bins.RAST | Genome | RAST annotation |
| CNT.126__Fire_bins.RAST | Genome | RAST annotation |
| CNT.345__Fire_bins.RAST | Genome | RAST annotation |
| CLV.161__Fire_bins.RAST | Genome | RAST annotation |
| KNG.820__Fire_bins.RAST | Genome | RAST annotation |
| KNG.331__Fire_bins.RAST | Genome | RAST annotation |
| KNG.728__Fire_bins.RAST | Genome | RAST annotation |
| KNG.226__Fire_bins.RAST | Genome | RAST annotation |
| KNG.058__Fire_bins.RAST | Genome | RAST annotation |
| KNG.262__Fire_bins.RAST | Genome | RAST annotation |
| FDR.560__Fire_bins.RAST | Genome | RAST annotation |
| CNT.333__Fire_bins.RAST | Genome | RAST annotation |
| FDR.338__Fire_bins.RAST | Genome | RAST annotation |
| KNG.186__Fire_bins.RAST | Genome | RAST annotation |
| CNT.193__Fire_bins.RAST | Genome | RAST annotation |
| FDR.010__Fire_bins.RAST | Genome | RAST annotation |
| CLV.378__Fire_bins.RAST | Genome | RAST annotation |
| KNG.761__Fire_bins.RAST | Genome | RAST annotation |
| CNT.079__Fire_bins.RAST | Genome | RAST annotation |
| KNG.610__Fire_bins.RAST | Genome | RAST annotation |
| KNG.802__Fire_bins.RAST | Genome | RAST annotation |
| CLV.169__Fire_bins.RAST | Genome | RAST annotation |
| KNG.302__Fire_bins.RAST | Genome | RAST annotation |
| FDR.444__Fire_bins.RAST | Genome | RAST annotation |
| KNG.120__Fire_bins.RAST | Genome | RAST annotation |
| CNT.289__Fire_bins.RAST | Genome | RAST annotation |
| KNG.009__Fire_bins.RAST | Genome | RAST annotation |
| CLV.099__Fire_bins.RAST | Genome | RAST annotation |
| FDR.112__Fire_bins.RAST | Genome | RAST annotation |
| FDR.522__Fire_bins.RAST | Genome | RAST annotation |
| CNT.137__Fire_bins.RAST | Genome | RAST annotation |
| CLV.310__Fire_bins.RAST | Genome | RAST annotation |
| KNG.316__Fire_bins.RAST | Genome | RAST annotation |
| KNG.699__Fire_bins.RAST | Genome | RAST annotation |
| KNG.408__Fire_bins.RAST | Genome | RAST annotation |
| KNG.006__Fire_bins.RAST | Genome | RAST annotation |
| KNG.216__Fire_bins.RAST | Genome | RAST annotation |
| KNG.044__Fire_bins.RAST | Genome | RAST annotation |
| CNT.186__Fire_bins.RAST | Genome | RAST annotation |
| CNT.536__Fire_bins.RAST | Genome | RAST annotation |
| KNG.833__Fire_bins.RAST | Genome | RAST annotation |
| FDR.359__Fire_bins.RAST | Genome | RAST annotation |
| CNT.212__Fire_bins.RAST | Genome | RAST annotation |
| CNT.091__Fire_bins.RAST | Genome | RAST annotation |
| KNG.578__Fire_bins.RAST | Genome | RAST annotation |
| KNG.794__Fire_bins.RAST | Genome | RAST annotation |
| CLV.115__Fire_bins.RAST | Genome | RAST annotation |
| KNG.020__Fire_bins.RAST | Genome | RAST annotation |
| FDR.614__Fire_bins.RAST | Genome | RAST annotation |
| KNG.305__Fire_bins.RAST | Genome | RAST annotation |
| KNG.146__Fire_bins.RAST | Genome | RAST annotation |
| FDR.101__Fire_bins.RAST | Genome | RAST annotation |
| CNT.284__Fire_bins.RAST | Genome | RAST annotation |
| KNG.281__Fire_bins.RAST | Genome | RAST annotation |
| FDR.521__Fire_bins.RAST | Genome | RAST annotation |
| CNT.134__Fire_bins.RAST | Genome | RAST annotation |
| CNT.405__Fire_bins.RAST | Genome | RAST annotation |
| CLV.266__Fire_bins.RAST | Genome | RAST annotation |
| KNG.322__Fire_bins.RAST | Genome | RAST annotation |
| KNG.710__Fire_bins.RAST | Genome | RAST annotation |
| FDR.628__Fire_bins.RAST | Genome | RAST annotation |
| KNG.421__Fire_bins.RAST | Genome | RAST annotation |
| KNG.046__Fire_bins.RAST | Genome | RAST annotation |
| KNG.219__Fire_bins.RAST | Genome | RAST annotation |
| FDR.480__Fire_bins.RAST | Genome | RAST annotation |
| FDR.554__Fire_bins.RAST | Genome | RAST annotation |
| CNT.531__Fire_bins.RAST | Genome | RAST annotation |
| CNT.279__Fire_bins.RAST | Genome | RAST annotation |
| CNT.025__Fire_bins.RAST | Genome | RAST annotation |
| KNG.514__Fire_bins.RAST | Genome | RAST annotation |
| KNG.799__Fire_bins.RAST | Genome | RAST annotation |
| CLV.104__Fire_bins.RAST | Genome | RAST annotation |
| FDR.489__Fire_bins.RAST | Genome | RAST annotation |
| KNG.108__Fire_bins.RAST | Genome | RAST annotation |
| KNG.252__Fire_bins.RAST | Genome | RAST annotation |
| CLV.081__Fire_bins.RAST | Genome | RAST annotation |
| CNT.110__Fire_bins.RAST | Genome | RAST annotation |
| CNT.308__Fire_bins.RAST | Genome | RAST annotation |
| FDR.178__Fire_bins.RAST | Genome | RAST annotation |
| CNT.056__Fire_bins.RAST | Genome | RAST annotation |
| CLV.316__Fire_bins.RAST | Genome | RAST annotation |
| KNG.829__Fire_bins.RAST | Genome | RAST annotation |
| KNG.670__Fire_bins.RAST | Genome | RAST annotation |
| KNG.392__Fire_bins.RAST | Genome | RAST annotation |
| KNG.005__Fire_bins.RAST | Genome | RAST annotation |
| KNG.085__Fire_bins.RAST | Genome | RAST annotation |
| KNG.747__Fire_bins.RAST | Genome | RAST annotation |
| FDR.226__Fire_bins.RAST | Genome | RAST annotation |
| KNG.195__Fire_bins.RAST | Genome | RAST annotation |
| KNG.027__Fire_bins.RAST | Genome | RAST annotation |
| CNT.191__Fire_bins.RAST | Genome | RAST annotation |
| FDR.001__Fire_bins.RAST | Genome | RAST annotation |
| FDR.416__Fire_bins.RAST | Genome | RAST annotation |
| KNG.830__Fire_bins.RAST | Genome | RAST annotation |
| CNT.036__Fire_bins.RAST | Genome | RAST annotation |
| CNT.161__Fire_bins.RAST | Genome | RAST annotation |
| KNG.448__Fire_bins.RAST | Genome | RAST annotation |
| KNG.813__Fire_bins.RAST | Genome | RAST annotation |
| KNG.014__Fire_bins.RAST | Genome | RAST annotation |
| KNG.566__Fire_bins.RAST | Genome | RAST annotation |
| FDR.445__Fire_bins.RAST | Genome | RAST annotation |
| KNG.259__Fire_bins.RAST | Genome | RAST annotation |
| KNG.118__Fire_bins.RAST | Genome | RAST annotation |
| CLV.082__Fire_bins.RAST | Genome | RAST annotation |
| CNT.105__Fire_bins.RAST | Genome | RAST annotation |
| FDR.620__Fire_bins.RAST | Genome | RAST annotation |
| FDR.549__Fire_bins.RAST | Genome | RAST annotation |
| CNT.293__Fire_bins.RAST | Genome | RAST annotation |
| FDR.171__Fire_bins.RAST | Genome | RAST annotation |
| CNT.150__Fire_bins.RAST | Genome | RAST annotation |
| CLV.312__Fire_bins.RAST | Genome | RAST annotation |
| KNG.688__Fire_bins.RAST | Genome | RAST annotation |
| KNG.402__Fire_bins.RAST | Genome | RAST annotation |
| FDR.625__Fire_bins.RAST | Genome | RAST annotation |
| KNG.033__Fire_bins.RAST | Genome | RAST annotation |
| KNG.210__Fire_bins.RAST | Genome | RAST annotation |
| CNT.188__Fire_bins.RAST | Genome | RAST annotation |
| CNT.447__Fire_bins.RAST | Genome | RAST annotation |
| KNG.832__Fire_bins.RAST | Genome | RAST annotation |
| FDR.407__Fire_bins.RAST | Genome | RAST annotation |
| CNT.271__Fire_bins.RAST | Genome | RAST annotation |
| CNT.029__Fire_bins.RAST | Genome | RAST annotation |
| KNG.571__Fire_bins.RAST | Genome | RAST annotation |
| KNG.793__Fire_bins.RAST | Genome | RAST annotation |
| KNG.645__Fire_bins.RAST | Genome | RAST annotation |
| KNG.015__Fire_bins.RAST | Genome | RAST annotation |
| KNG.012__Fire_bins.RAST | Genome | RAST annotation |
| KNG.135__Fire_bins.RAST | Genome | RAST annotation |
| KNG.247__Fire_bins.RAST | Genome | RAST annotation |
| FDR.048__Fire_bins.RAST | Genome | RAST annotation |
| FDR.501__Fire_bins.RAST | Genome | RAST annotation |
| CNT.124__Fire_bins.RAST | Genome | RAST annotation |
| CNT.331__Fire_bins.RAST | Genome | RAST annotation |
| FDR.211__Fire_bins.RAST | Genome | RAST annotation |
| CLV.173__Fire_bins.RAST | Genome | RAST annotation |
| KNG.826__Fire_bins.RAST | Genome | RAST annotation |
| KNG.339__Fire_bins.RAST | Genome | RAST annotation |
| KNG.732__Fire_bins.RAST | Genome | RAST annotation |
| KNG.080__Fire_bins.RAST | Genome | RAST annotation |
| KNG.061__Fire_bins.RAST | Genome | RAST annotation |
| FDR.595__Fire_bins.RAST | Genome | RAST annotation |
| FDR.312__Fire_bins.RAST | Genome | RAST annotation |
| KNG.190__Fire_bins.RAST | Genome | RAST annotation |
| CNT.470__Fire_bins.RAST | Genome | RAST annotation |
| CNT.192__Fire_bins.RAST | Genome | RAST annotation |
| CLV.325__Fire_bins.RAST | Genome | RAST annotation |
| KNG.766__Fire_bins.RAST | Genome | RAST annotation |
| CNT.016__Fire_bins.RAST | Genome | RAST annotation |
| KNG.540__Fire_bins.RAST | Genome | RAST annotation |
| KNG.807__Fire_bins.RAST | Genome | RAST annotation |
| KNG.178__Fire_bins.RAST | Genome | RAST annotation |
| FDR.018__Fire_bins.RAST | Genome | RAST annotation |
| CLV.031__Fire_bins.RAST | Genome | RAST annotation |
| KNG.299__Fire_bins.RAST | Genome | RAST annotation |
| KNG.140__Fire_bins.RAST | Genome | RAST annotation |
| FDR.500__Fire_bins.RAST | Genome | RAST annotation |
| CNT.121__Fire_bins.RAST | Genome | RAST annotation |
| FDR.187__Fire_bins.RAST | Genome | RAST annotation |
| CNT.312__Fire_bins.RAST | Genome | RAST annotation |
| CNT.097__Fire_bins.RAST | Genome | RAST annotation |
| KNG.653__Fire_bins.RAST | Genome | RAST annotation |
| KNG.828__Fire_bins.RAST | Genome | RAST annotation |
| KNG.348__Fire_bins.RAST | Genome | RAST annotation |
| KNG.740__Fire_bins.RAST | Genome | RAST annotation |
| FDR.606__Fire_bins.RAST | Genome | RAST annotation |
| KNG.083__Fire_bins.RAST | Genome | RAST annotation |
| FDR.307__Fire_bins.RAST | Genome | RAST annotation |
| FDR.566__Fire_bins.RAST | Genome | RAST annotation |
| KNG.192__Fire_bins.RAST | Genome | RAST annotation |
| CNT.275__Fire_bins.RAST | Genome | RAST annotation |
| FDR.436__Fire_bins.RAST | Genome | RAST annotation |
| CNT.474__Fire_bins.RAST | Genome | RAST annotation |
| CLV.332__Fire_bins.RAST | Genome | RAST annotation |
| KNG.774__Fire_bins.RAST | Genome | RAST annotation |
| CNT.009__Fire_bins.RAST | Genome | RAST annotation |
| KNG.559__Fire_bins.RAST | Genome | RAST annotation |
| KNG.013__Fire_bins.RAST | Genome | RAST annotation |
| KNG.177__Fire_bins.RAST | Genome | RAST annotation |
| Fire_genomes | GenomeSet | Genome Set |
Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 816 contigs containing 2011817 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2867 new features were called, of which 223 are non-coding. Output genome has the following feature types: Coding gene 2644 Non-coding repeat 196 Non-coding rna 27 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.011__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 403 contigs containing 5556636 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6238 new features were called, of which 289 are non-coding. Output genome has the following feature types: Coding gene 5949 Non-coding repeat 257 Non-coding rna 32 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.159__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 317 contigs containing 3143653 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3504 new features were called, of which 149 are non-coding. Output genome has the following feature types: Coding gene 3355 Non-coding repeat 104 Non-coding rna 45 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.307__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 3196 contigs containing 5839152 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8380 new features were called, of which 315 are non-coding. Output genome has the following feature types: Coding gene 8065 Non-coding repeat 305 Non-coding rna 10 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.259__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1080 contigs containing 4017472 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4407 new features were called, of which 41 are non-coding. Output genome has the following feature types: Coding gene 4366 Non-coding repeat 21 Non-coding rna 20 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.075__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1010 contigs containing 1722310 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2799 new features were called, of which 33 are non-coding. Output genome has the following feature types: Coding gene 2766 Non-coding repeat 12 Non-coding rna 21 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.512__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1257 contigs containing 4460235 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5672 new features were called, of which 716 are non-coding. Output genome has the following feature types: Coding gene 4956 Non-coding repeat 679 Non-coding rna 37 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.284__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 884 contigs containing 4929573 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6142 new features were called, of which 656 are non-coding. Output genome has the following feature types: Coding gene 5486 Non-coding repeat 581 Non-coding rna 75 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.132__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 737 contigs containing 4170599 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4673 new features were called, of which 200 are non-coding. Output genome has the following feature types: Coding gene 4473 Non-coding repeat 168 Non-coding rna 32 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.381__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2305 contigs containing 4041584 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7033 new features were called, of which 1074 are non-coding. Output genome has the following feature types: Coding gene 5959 Non-coding repeat 1054 Non-coding rna 20 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.236__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1238 contigs containing 4280471 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5759 new features were called, of which 677 are non-coding. Output genome has the following feature types: Coding gene 5082 Non-coding repeat 643 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.324__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1017 contigs containing 6021123 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6927 new features were called, of which 551 are non-coding. Output genome has the following feature types: Coding gene 6376 Non-coding repeat 507 Non-coding rna 44 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.716__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 441 contigs containing 3251293 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3883 new features were called, of which 423 are non-coding. Output genome has the following feature types: Coding gene 3460 Non-coding repeat 400 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.225__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1069 contigs containing 9613772 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 10436 new features were called, of which 561 are non-coding. Output genome has the following feature types: Coding gene 9875 Non-coding repeat 522 Non-coding rna 39 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.556__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 461 contigs containing 5072918 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5500 new features were called, of which 584 are non-coding. Output genome has the following feature types: Coding gene 4916 Non-coding repeat 544 Non-coding rna 40 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.051__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1695 contigs containing 5065668 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7283 new features were called, of which 820 are non-coding. Output genome has the following feature types: Coding gene 6463 Non-coding repeat 795 Non-coding rna 25 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.348__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1300 contigs containing 3884696 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4939 new features were called, of which 343 are non-coding. Output genome has the following feature types: Coding gene 4596 Non-coding repeat 314 Non-coding rna 29 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.435__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1893 contigs containing 4536215 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6887 new features were called, of which 969 are non-coding. Output genome has the following feature types: Coding gene 5918 Non-coding repeat 951 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.195__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 110 contigs containing 4923164 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4827 new features were called, of which 219 are non-coding. Output genome has the following feature types: Coding gene 4608 Non-coding repeat 175 Non-coding rna 44 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.528__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1258 contigs containing 4975503 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7125 new features were called, of which 1249 are non-coding. Output genome has the following feature types: Coding gene 5876 Non-coding repeat 1210 Non-coding rna 39 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.089__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 656 contigs containing 3430679 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4212 new features were called, of which 271 are non-coding. Output genome has the following feature types: Coding gene 3941 Non-coding repeat 238 Non-coding rna 33 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.534__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 808 contigs containing 3913105 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5095 new features were called, of which 560 are non-coding. Output genome has the following feature types: Coding gene 4535 Non-coding repeat 526 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.800__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 649 contigs containing 4673400 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5315 new features were called, of which 438 are non-coding. Output genome has the following feature types: Coding gene 4877 Non-coding repeat 374 Non-coding rna 64 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.153__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 246 contigs containing 3176129 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3239 new features were called, of which 108 are non-coding. Output genome has the following feature types: Coding gene 3131 Non-coding repeat 76 Non-coding rna 32 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.165__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2516 contigs containing 5082334 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7879 new features were called, of which 1068 are non-coding. Output genome has the following feature types: Coding gene 6811 Non-coding repeat 1037 Non-coding rna 31 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.003__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 876 contigs containing 3404624 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4768 new features were called, of which 878 are non-coding. Output genome has the following feature types: Coding gene 3890 Non-coding repeat 836 Non-coding rna 42 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.502__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 973 contigs containing 4941613 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6591 new features were called, of which 498 are non-coding. Output genome has the following feature types: Coding gene 6093 Non-coding repeat 449 Non-coding rna 49 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.073__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 787 contigs containing 4508054 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5845 new features were called, of which 685 are non-coding. Output genome has the following feature types: Coding gene 5160 Non-coding repeat 651 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.244__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 483 contigs containing 5007353 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5463 new features were called, of which 309 are non-coding. Output genome has the following feature types: Coding gene 5154 Non-coding repeat 263 Non-coding rna 46 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.126__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1564 contigs containing 6560917 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7566 new features were called, of which 383 are non-coding. Output genome has the following feature types: Coding gene 7183 Non-coding repeat 360 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.345__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 226 contigs containing 6472807 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6133 new features were called, of which 329 are non-coding. Output genome has the following feature types: Coding gene 5804 Non-coding repeat 259 Non-coding rna 70 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.161__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 495 contigs containing 1210938 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 1828 new features were called, of which 63 are non-coding. Output genome has the following feature types: Coding gene 1765 Non-coding repeat 11 Non-coding rna 52 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.820__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 807 contigs containing 6871504 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8294 new features were called, of which 833 are non-coding. Output genome has the following feature types: Coding gene 7461 Non-coding repeat 773 Non-coding rna 60 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.331__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1055 contigs containing 3556553 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4774 new features were called, of which 801 are non-coding. Output genome has the following feature types: Coding gene 3973 Non-coding repeat 783 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.728__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 962 contigs containing 3668494 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4735 new features were called, of which 602 are non-coding. Output genome has the following feature types: Coding gene 4133 Non-coding repeat 561 Non-coding rna 41 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.226__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2099 contigs containing 8107791 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 10872 new features were called, of which 1853 are non-coding. Output genome has the following feature types: Coding gene 9019 Non-coding repeat 1816 Non-coding rna 37 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.058__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1969 contigs containing 8167303 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 11312 new features were called, of which 2072 are non-coding. Output genome has the following feature types: Coding gene 9240 Non-coding repeat 2023 Non-coding rna 49 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.262__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 653 contigs containing 6141060 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6542 new features were called, of which 731 are non-coding. Output genome has the following feature types: Coding gene 5811 Non-coding repeat 695 Non-coding rna 36 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.560__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 833 contigs containing 4810998 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6425 new features were called, of which 1103 are non-coding. Output genome has the following feature types: Coding gene 5322 Non-coding repeat 1056 Non-coding rna 47 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.333__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1434 contigs containing 4122592 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5544 new features were called, of which 453 are non-coding. Output genome has the following feature types: Coding gene 5091 Non-coding repeat 436 Non-coding rna 17 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.338__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1721 contigs containing 8739110 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 10221 new features were called, of which 484 are non-coding. Output genome has the following feature types: Coding gene 9737 Non-coding repeat 448 Non-coding rna 36 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.186__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 394 contigs containing 3594361 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4292 new features were called, of which 298 are non-coding. Output genome has the following feature types: Coding gene 3994 Non-coding repeat 252 Non-coding rna 46 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.193__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 884 contigs containing 3726294 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4999 new features were called, of which 551 are non-coding. Output genome has the following feature types: Coding gene 4448 Non-coding repeat 524 Non-coding rna 27 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.010__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1916 contigs containing 5608070 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8181 new features were called, of which 1002 are non-coding. Output genome has the following feature types: Coding gene 7179 Non-coding repeat 966 Non-coding rna 36 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.378__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1595 contigs containing 2369457 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3637 new features were called, of which 133 are non-coding. Output genome has the following feature types: Coding gene 3504 Non-coding repeat 122 Non-coding rna 11 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.761__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1444 contigs containing 4134306 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5567 new features were called, of which 856 are non-coding. Output genome has the following feature types: Coding gene 4711 Non-coding repeat 838 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.079__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1385 contigs containing 3980601 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5264 new features were called, of which 799 are non-coding. Output genome has the following feature types: Coding gene 4465 Non-coding repeat 777 Non-coding rna 22 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.610__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 646 contigs containing 5047851 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5630 new features were called, of which 613 are non-coding. Output genome has the following feature types: Coding gene 5017 Non-coding repeat 576 Non-coding rna 37 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.802__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 226 contigs containing 2541899 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2854 new features were called, of which 113 are non-coding. Output genome has the following feature types: Coding gene 2741 Non-coding repeat 72 Non-coding rna 41 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.169__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 417 contigs containing 4910571 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5355 new features were called, of which 340 are non-coding. Output genome has the following feature types: Coding gene 5015 Non-coding repeat 299 Non-coding rna 41 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.302__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1193 contigs containing 3837815 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5419 new features were called, of which 178 are non-coding. Output genome has the following feature types: Coding gene 5241 Non-coding repeat 149 Non-coding rna 29 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.444__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 581 contigs containing 6421207 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6843 new features were called, of which 760 are non-coding. Output genome has the following feature types: Coding gene 6083 Non-coding repeat 717 Non-coding rna 43 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.120__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1721 contigs containing 4410408 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6797 new features were called, of which 1012 are non-coding. Output genome has the following feature types: Coding gene 5785 Non-coding repeat 987 Non-coding rna 25 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.289__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 488 contigs containing 3528084 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4152 new features were called, of which 334 are non-coding. Output genome has the following feature types: Coding gene 3818 Non-coding repeat 305 Non-coding rna 29 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.009__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2744 contigs containing 7089368 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 9247 new features were called, of which 687 are non-coding. Output genome has the following feature types: Coding gene 8560 Non-coding repeat 664 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.099__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1305 contigs containing 3729084 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5319 new features were called, of which 754 are non-coding. Output genome has the following feature types: Coding gene 4565 Non-coding repeat 739 Non-coding rna 15 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.112__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 503 contigs containing 5355603 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6123 new features were called, of which 639 are non-coding. Output genome has the following feature types: Coding gene 5484 Non-coding repeat 581 Non-coding rna 58 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.522__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 993 contigs containing 2863173 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3906 new features were called, of which 555 are non-coding. Output genome has the following feature types: Coding gene 3351 Non-coding repeat 532 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.137__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1019 contigs containing 3147370 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4551 new features were called, of which 484 are non-coding. Output genome has the following feature types: Coding gene 4067 Non-coding repeat 463 Non-coding rna 21 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.310__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 108 contigs containing 1025125 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 1298 new features were called, of which 127 are non-coding. Output genome has the following feature types: Coding gene 1171 Non-coding repeat 86 Non-coding rna 41 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.316__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1049 contigs containing 5175235 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6047 new features were called, of which 603 are non-coding. Output genome has the following feature types: Coding gene 5444 Non-coding repeat 537 Non-coding rna 66 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.699__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 476 contigs containing 2383671 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2784 new features were called, of which 142 are non-coding. Output genome has the following feature types: Coding gene 2642 Non-coding repeat 124 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.408__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2235 contigs containing 3508336 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6057 new features were called, of which 708 are non-coding. Output genome has the following feature types: Coding gene 5349 Non-coding repeat 692 Non-coding rna 16 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.006__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1366 contigs containing 4562984 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6440 new features were called, of which 1483 are non-coding. Output genome has the following feature types: Coding gene 4957 Non-coding repeat 1445 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.216__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 574 contigs containing 2369465 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3041 new features were called, of which 325 are non-coding. Output genome has the following feature types: Coding gene 2716 Non-coding repeat 293 Non-coding rna 32 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.044__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1965 contigs containing 4479639 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5997 new features were called, of which 422 are non-coding. Output genome has the following feature types: Coding gene 5575 Non-coding repeat 404 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.186__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 792 contigs containing 4209544 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4828 new features were called, of which 92 are non-coding. Output genome has the following feature types: Coding gene 4736 Non-coding repeat 50 Non-coding rna 42 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.536__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 776 contigs containing 4934530 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6515 new features were called, of which 967 are non-coding. Output genome has the following feature types: Coding gene 5548 Non-coding repeat 890 Non-coding rna 77 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.833__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1385 contigs containing 6352811 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8163 new features were called, of which 1084 are non-coding. Output genome has the following feature types: Coding gene 7079 Non-coding repeat 1053 Non-coding rna 31 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.359__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2441 contigs containing 7634744 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 9052 new features were called, of which 256 are non-coding. Output genome has the following feature types: Coding gene 8796 Non-coding repeat 208 Non-coding rna 48 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.212__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 991 contigs containing 4177674 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5530 new features were called, of which 170 are non-coding. Output genome has the following feature types: Coding gene 5360 Non-coding repeat 126 Non-coding rna 44 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.091__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2647 contigs containing 3862298 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5543 new features were called, of which 27 are non-coding. Output genome has the following feature types: Coding gene 5516 Non-coding repeat 10 Non-coding rna 17 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.578__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2189 contigs containing 5760379 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8464 new features were called, of which 1680 are non-coding. Output genome has the following feature types: Coding gene 6784 Non-coding repeat 1663 Non-coding rna 17 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.794__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 789 contigs containing 3067474 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4305 new features were called, of which 592 are non-coding. Output genome has the following feature types: Coding gene 3713 Non-coding repeat 557 Non-coding rna 35 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.115__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1700 contigs containing 3472515 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5916 new features were called, of which 1163 are non-coding. Output genome has the following feature types: Coding gene 4753 Non-coding repeat 1144 Non-coding rna 19 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.020__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1195 contigs containing 4339590 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5097 new features were called, of which 75 are non-coding. Output genome has the following feature types: Coding gene 5022 Non-coding repeat 47 Non-coding rna 28 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.614__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 377 contigs containing 903574 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 1212 new features were called, of which 72 are non-coding. Output genome has the following feature types: Coding gene 1140 Non-coding repeat 60 Non-coding rna 12 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.305__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 374 contigs containing 2830949 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3226 new features were called, of which 202 are non-coding. Output genome has the following feature types: Coding gene 3024 Non-coding repeat 167 Non-coding rna 35 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.146__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1560 contigs containing 4014529 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5224 new features were called, of which 201 are non-coding. Output genome has the following feature types: Coding gene 5023 Non-coding repeat 178 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.101__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1760 contigs containing 4982811 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6104 new features were called, of which 73 are non-coding. Output genome has the following feature types: Coding gene 6031 Non-coding repeat 40 Non-coding rna 33 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.284__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 193 contigs containing 4090211 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4277 new features were called, of which 179 are non-coding. Output genome has the following feature types: Coding gene 4098 Non-coding repeat 145 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.281__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 275 contigs containing 1408008 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 1753 new features were called, of which 114 are non-coding. Output genome has the following feature types: Coding gene 1639 Non-coding repeat 97 Non-coding rna 17 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.521__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 557 contigs containing 2265601 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3033 new features were called, of which 372 are non-coding. Output genome has the following feature types: Coding gene 2661 Non-coding repeat 354 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.134__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 195 contigs containing 7862211 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7563 new features were called, of which 251 are non-coding. Output genome has the following feature types: Coding gene 7312 Non-coding repeat 188 Non-coding rna 63 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.405__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1016 contigs containing 3931658 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5494 new features were called, of which 853 are non-coding. Output genome has the following feature types: Coding gene 4641 Non-coding repeat 832 Non-coding rna 21 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.266__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 845 contigs containing 3291576 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4028 new features were called, of which 381 are non-coding. Output genome has the following feature types: Coding gene 3647 Non-coding repeat 357 Non-coding rna 24 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.322__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1156 contigs containing 5023562 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7329 new features were called, of which 1207 are non-coding. Output genome has the following feature types: Coding gene 6122 Non-coding repeat 1157 Non-coding rna 50 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.710__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1355 contigs containing 3809476 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4931 new features were called, of which 291 are non-coding. Output genome has the following feature types: Coding gene 4640 Non-coding repeat 254 Non-coding rna 37 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.628__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 444 contigs containing 4137747 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4652 new features were called, of which 240 are non-coding. Output genome has the following feature types: Coding gene 4412 Non-coding repeat 194 Non-coding rna 46 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.421__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1809 contigs containing 6943696 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 9676 new features were called, of which 1178 are non-coding. Output genome has the following feature types: Coding gene 8498 Non-coding repeat 1148 Non-coding rna 30 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.046__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 588 contigs containing 2555988 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3259 new features were called, of which 260 are non-coding. Output genome has the following feature types: Coding gene 2999 Non-coding repeat 233 Non-coding rna 27 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.219__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1070 contigs containing 2412442 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3610 new features were called, of which 81 are non-coding. Output genome has the following feature types: Coding gene 3529 Non-coding repeat 48 Non-coding rna 33 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.480__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1449 contigs containing 4880249 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7072 new features were called, of which 598 are non-coding. Output genome has the following feature types: Coding gene 6474 Non-coding repeat 547 Non-coding rna 51 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.554__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 418 contigs containing 4447656 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4068 new features were called, of which 128 are non-coding. Output genome has the following feature types: Coding gene 3940 Non-coding repeat 116 Non-coding rna 12 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.531__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 914 contigs containing 3407656 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4411 new features were called, of which 404 are non-coding. Output genome has the following feature types: Coding gene 4007 Non-coding repeat 381 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.279__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 715 contigs containing 3085482 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4132 new features were called, of which 697 are non-coding. Output genome has the following feature types: Coding gene 3435 Non-coding repeat 654 Non-coding rna 43 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.025__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 987 contigs containing 5591022 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6981 new features were called, of which 686 are non-coding. Output genome has the following feature types: Coding gene 6295 Non-coding repeat 654 Non-coding rna 32 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.514__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 971 contigs containing 4548659 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5774 new features were called, of which 411 are non-coding. Output genome has the following feature types: Coding gene 5363 Non-coding repeat 360 Non-coding rna 51 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.799__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 370 contigs containing 3888801 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3878 new features were called, of which 202 are non-coding. Output genome has the following feature types: Coding gene 3676 Non-coding repeat 154 Non-coding rna 48 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.104__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 951 contigs containing 3263681 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4285 new features were called, of which 464 are non-coding. Output genome has the following feature types: Coding gene 3821 Non-coding repeat 445 Non-coding rna 19 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.489__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1951 contigs containing 8885523 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 11132 new features were called, of which 1412 are non-coding. Output genome has the following feature types: Coding gene 9720 Non-coding repeat 1384 Non-coding rna 28 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.108__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 640 contigs containing 3638581 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4402 new features were called, of which 488 are non-coding. Output genome has the following feature types: Coding gene 3914 Non-coding repeat 459 Non-coding rna 29 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.252__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1575 contigs containing 4498944 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6763 new features were called, of which 944 are non-coding. Output genome has the following feature types: Coding gene 5819 Non-coding repeat 908 Non-coding rna 36 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.081__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 756 contigs containing 3172284 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3995 new features were called, of which 504 are non-coding. Output genome has the following feature types: Coding gene 3491 Non-coding repeat 486 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.110__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 991 contigs containing 4797220 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6545 new features were called, of which 511 are non-coding. Output genome has the following feature types: Coding gene 6034 Non-coding repeat 464 Non-coding rna 47 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.308__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1637 contigs containing 3318643 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4434 new features were called, of which 321 are non-coding. Output genome has the following feature types: Coding gene 4113 Non-coding repeat 311 Non-coding rna 10 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.178__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 624 contigs containing 3603336 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4341 new features were called, of which 399 are non-coding. Output genome has the following feature types: Coding gene 3942 Non-coding repeat 365 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.056__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1795 contigs containing 4261040 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5875 new features were called, of which 313 are non-coding. Output genome has the following feature types: Coding gene 5562 Non-coding repeat 302 Non-coding rna 11 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.316__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 729 contigs containing 4747054 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5950 new features were called, of which 556 are non-coding. Output genome has the following feature types: Coding gene 5394 Non-coding repeat 485 Non-coding rna 71 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.829__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 918 contigs containing 6135782 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6620 new features were called, of which 491 are non-coding. Output genome has the following feature types: Coding gene 6129 Non-coding repeat 435 Non-coding rna 56 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.670__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1240 contigs containing 3598527 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4798 new features were called, of which 517 are non-coding. Output genome has the following feature types: Coding gene 4281 Non-coding repeat 495 Non-coding rna 22 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.392__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1781 contigs containing 8425574 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 10471 new features were called, of which 1266 are non-coding. Output genome has the following feature types: Coding gene 9205 Non-coding repeat 1227 Non-coding rna 39 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.005__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 353 contigs containing 3755523 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4274 new features were called, of which 350 are non-coding. Output genome has the following feature types: Coding gene 3924 Non-coding repeat 306 Non-coding rna 44 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.085__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 607 contigs containing 4126349 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4844 new features were called, of which 481 are non-coding. Output genome has the following feature types: Coding gene 4363 Non-coding repeat 449 Non-coding rna 32 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.747__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 586 contigs containing 4948261 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5663 new features were called, of which 291 are non-coding. Output genome has the following feature types: Coding gene 5372 Non-coding repeat 257 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.226__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1871 contigs containing 6524771 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 9323 new features were called, of which 1219 are non-coding. Output genome has the following feature types: Coding gene 8104 Non-coding repeat 1146 Non-coding rna 73 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.195__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1655 contigs containing 5488991 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8989 new features were called, of which 1474 are non-coding. Output genome has the following feature types: Coding gene 7515 Non-coding repeat 1422 Non-coding rna 52 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.027__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2169 contigs containing 4158871 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7166 new features were called, of which 1441 are non-coding. Output genome has the following feature types: Coding gene 5725 Non-coding repeat 1432 Non-coding rna 9 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.191__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1403 contigs containing 3298373 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4801 new features were called, of which 545 are non-coding. Output genome has the following feature types: Coding gene 4256 Non-coding repeat 522 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.001__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 374 contigs containing 3679818 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4112 new features were called, of which 227 are non-coding. Output genome has the following feature types: Coding gene 3885 Non-coding repeat 187 Non-coding rna 40 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.416__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 43 contigs containing 3499990 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3281 new features were called, of which 51 are non-coding. Output genome has the following feature types: Coding gene 3230 Non-coding repeat 16 Non-coding rna 35 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.830__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 416 contigs containing 3624942 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4156 new features were called, of which 213 are non-coding. Output genome has the following feature types: Coding gene 3943 Non-coding repeat 183 Non-coding rna 30 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.036__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 595 contigs containing 5223083 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5334 new features were called, of which 128 are non-coding. Output genome has the following feature types: Coding gene 5206 Non-coding repeat 84 Non-coding rna 44 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.161__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1071 contigs containing 6365161 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8478 new features were called, of which 1059 are non-coding. Output genome has the following feature types: Coding gene 7419 Non-coding repeat 1016 Non-coding rna 43 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.448__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 799 contigs containing 5764910 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6854 new features were called, of which 487 are non-coding. Output genome has the following feature types: Coding gene 6367 Non-coding repeat 451 Non-coding rna 36 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.813__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1919 contigs containing 5509653 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8698 new features were called, of which 1572 are non-coding. Output genome has the following feature types: Coding gene 7126 Non-coding repeat 1534 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.014__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1483 contigs containing 5946735 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7727 new features were called, of which 1414 are non-coding. Output genome has the following feature types: Coding gene 6313 Non-coding repeat 1383 Non-coding rna 31 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.566__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1609 contigs containing 5877328 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7610 new features were called, of which 502 are non-coding. Output genome has the following feature types: Coding gene 7108 Non-coding repeat 462 Non-coding rna 40 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.445__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 474 contigs containing 3505030 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4417 new features were called, of which 558 are non-coding. Output genome has the following feature types: Coding gene 3859 Non-coding repeat 521 Non-coding rna 37 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.259__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 834 contigs containing 5499056 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6541 new features were called, of which 588 are non-coding. Output genome has the following feature types: Coding gene 5953 Non-coding repeat 552 Non-coding rna 36 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.118__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 193 contigs containing 6537038 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6596 new features were called, of which 323 are non-coding. Output genome has the following feature types: Coding gene 6273 Non-coding repeat 257 Non-coding rna 66 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.082__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1198 contigs containing 2824479 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3475 new features were called, of which 154 are non-coding. Output genome has the following feature types: Coding gene 3321 Non-coding repeat 142 Non-coding rna 12 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.105__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 176 contigs containing 4387317 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4346 new features were called, of which 116 are non-coding. Output genome has the following feature types: Coding gene 4230 Non-coding repeat 53 Non-coding rna 63 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.620__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2801 contigs containing 4780037 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6242 new features were called, of which 283 are non-coding. Output genome has the following feature types: Coding gene 5959 Non-coding repeat 266 Non-coding rna 17 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.549__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 169 contigs containing 6023477 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5858 new features were called, of which 314 are non-coding. Output genome has the following feature types: Coding gene 5544 Non-coding repeat 246 Non-coding rna 68 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.293__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1919 contigs containing 5046090 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8039 new features were called, of which 1608 are non-coding. Output genome has the following feature types: Coding gene 6431 Non-coding repeat 1586 Non-coding rna 22 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.171__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 568 contigs containing 3266544 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4191 new features were called, of which 696 are non-coding. Output genome has the following feature types: Coding gene 3495 Non-coding repeat 657 Non-coding rna 39 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.150__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1163 contigs containing 4296132 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4833 new features were called, of which 125 are non-coding. Output genome has the following feature types: Coding gene 4708 Non-coding repeat 100 Non-coding rna 25 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.312__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 548 contigs containing 7031355 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7487 new features were called, of which 397 are non-coding. Output genome has the following feature types: Coding gene 7090 Non-coding repeat 338 Non-coding rna 59 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.688__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1148 contigs containing 3324670 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4476 new features were called, of which 542 are non-coding. Output genome has the following feature types: Coding gene 3934 Non-coding repeat 524 Non-coding rna 18 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.402__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1271 contigs containing 3094571 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4064 new features were called, of which 214 are non-coding. Output genome has the following feature types: Coding gene 3850 Non-coding repeat 184 Non-coding rna 30 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.625__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 345 contigs containing 2804812 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3202 new features were called, of which 216 are non-coding. Output genome has the following feature types: Coding gene 2986 Non-coding repeat 182 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.033__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1303 contigs containing 5500385 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7597 new features were called, of which 1351 are non-coding. Output genome has the following feature types: Coding gene 6246 Non-coding repeat 1327 Non-coding rna 24 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.210__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 797 contigs containing 3019334 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3917 new features were called, of which 227 are non-coding. Output genome has the following feature types: Coding gene 3690 Non-coding repeat 217 Non-coding rna 10 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.188__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 351 contigs containing 4479608 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4952 new features were called, of which 313 are non-coding. Output genome has the following feature types: Coding gene 4639 Non-coding repeat 248 Non-coding rna 65 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.447__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1799 contigs containing 3158123 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4480 new features were called, of which 34 are non-coding. Output genome has the following feature types: Coding gene 4446 Non-coding repeat 13 Non-coding rna 21 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.832__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 661 contigs containing 4633147 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5908 new features were called, of which 890 are non-coding. Output genome has the following feature types: Coding gene 5018 Non-coding repeat 826 Non-coding rna 64 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.407__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 295 contigs containing 7909537 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7672 new features were called, of which 473 are non-coding. Output genome has the following feature types: Coding gene 7199 Non-coding repeat 422 Non-coding rna 51 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.271__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 782 contigs containing 4547996 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5748 new features were called, of which 514 are non-coding. Output genome has the following feature types: Coding gene 5234 Non-coding repeat 484 Non-coding rna 30 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.029__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 3218 contigs containing 5932640 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8670 new features were called, of which 269 are non-coding. Output genome has the following feature types: Coding gene 8401 Non-coding repeat 224 Non-coding rna 45 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.571__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 592 contigs containing 4245379 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5040 new features were called, of which 457 are non-coding. Output genome has the following feature types: Coding gene 4583 Non-coding repeat 398 Non-coding rna 59 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.793__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1520 contigs containing 3649170 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5963 new features were called, of which 1439 are non-coding. Output genome has the following feature types: Coding gene 4524 Non-coding repeat 1401 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.645__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1880 contigs containing 6153605 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8601 new features were called, of which 1121 are non-coding. Output genome has the following feature types: Coding gene 7480 Non-coding repeat 1083 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.015__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 677 contigs containing 4795173 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5717 new features were called, of which 403 are non-coding. Output genome has the following feature types: Coding gene 5314 Non-coding repeat 363 Non-coding rna 40 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.012__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1104 contigs containing 3901667 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5381 new features were called, of which 1018 are non-coding. Output genome has the following feature types: Coding gene 4363 Non-coding repeat 989 Non-coding rna 29 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.135__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 586 contigs containing 6000719 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6297 new features were called, of which 217 are non-coding. Output genome has the following feature types: Coding gene 6080 Non-coding repeat 172 Non-coding rna 45 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.247__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 569 contigs containing 4366221 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4957 new features were called, of which 275 are non-coding. Output genome has the following feature types: Coding gene 4682 Non-coding repeat 231 Non-coding rna 44 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.048__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1970 contigs containing 7324218 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 11766 new features were called, of which 2460 are non-coding. Output genome has the following feature types: Coding gene 9306 Non-coding repeat 2422 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.501__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 423 contigs containing 4669307 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4845 new features were called, of which 290 are non-coding. Output genome has the following feature types: Coding gene 4555 Non-coding repeat 247 Non-coding rna 43 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.124__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2149 contigs containing 5225806 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5832 new features were called, of which 147 are non-coding. Output genome has the following feature types: Coding gene 5685 Non-coding repeat 130 Non-coding rna 17 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.331__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 501 contigs containing 5312265 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5982 new features were called, of which 422 are non-coding. Output genome has the following feature types: Coding gene 5560 Non-coding repeat 352 Non-coding rna 70 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.211__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 256 contigs containing 5432048 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5419 new features were called, of which 166 are non-coding. Output genome has the following feature types: Coding gene 5253 Non-coding repeat 110 Non-coding rna 56 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.173__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 241 contigs containing 5939396 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6331 new features were called, of which 365 are non-coding. Output genome has the following feature types: Coding gene 5966 Non-coding repeat 316 Non-coding rna 49 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.826__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1063 contigs containing 3744360 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4677 new features were called, of which 68 are non-coding. Output genome has the following feature types: Coding gene 4609 Non-coding repeat 52 Non-coding rna 16 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.339__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 684 contigs containing 2862560 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3760 new features were called, of which 574 are non-coding. Output genome has the following feature types: Coding gene 3186 Non-coding repeat 539 Non-coding rna 35 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.732__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1778 contigs containing 4192059 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6765 new features were called, of which 1254 are non-coding. Output genome has the following feature types: Coding gene 5511 Non-coding repeat 1235 Non-coding rna 19 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.080__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1868 contigs containing 5227485 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8646 new features were called, of which 1986 are non-coding. Output genome has the following feature types: Coding gene 6660 Non-coding repeat 1964 Non-coding rna 22 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.061__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1248 contigs containing 5653251 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7537 new features were called, of which 826 are non-coding. Output genome has the following feature types: Coding gene 6711 Non-coding repeat 772 Non-coding rna 54 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.595__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 684 contigs containing 2022758 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2936 new features were called, of which 486 are non-coding. Output genome has the following feature types: Coding gene 2450 Non-coding repeat 455 Non-coding rna 31 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.312__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1619 contigs containing 4129757 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6717 new features were called, of which 1569 are non-coding. Output genome has the following feature types: Coding gene 5148 Non-coding repeat 1531 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.190__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 813 contigs containing 2613832 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3667 new features were called, of which 389 are non-coding. Output genome has the following feature types: Coding gene 3278 Non-coding repeat 359 Non-coding rna 30 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.470__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1198 contigs containing 4754656 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6418 new features were called, of which 289 are non-coding. Output genome has the following feature types: Coding gene 6129 Non-coding repeat 244 Non-coding rna 45 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.192__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1840 contigs containing 3592105 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5873 new features were called, of which 613 are non-coding. Output genome has the following feature types: Coding gene 5260 Non-coding repeat 583 Non-coding rna 30 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.325__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 863 contigs containing 3546834 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4689 new features were called, of which 518 are non-coding. Output genome has the following feature types: Coding gene 4171 Non-coding repeat 465 Non-coding rna 53 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.766__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1245 contigs containing 2934743 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4647 new features were called, of which 876 are non-coding. Output genome has the following feature types: Coding gene 3771 Non-coding repeat 852 Non-coding rna 24 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.016__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2297 contigs containing 4406373 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6331 new features were called, of which 788 are non-coding. Output genome has the following feature types: Coding gene 5543 Non-coding repeat 767 Non-coding rna 21 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.540__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 21 contigs containing 1126171 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 1253 new features were called, of which 68 are non-coding. Output genome has the following feature types: Coding gene 1185 Non-coding repeat 18 Non-coding rna 50 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.807__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1630 contigs containing 4130317 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6490 new features were called, of which 1266 are non-coding. Output genome has the following feature types: Coding gene 5224 Non-coding repeat 1228 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.178__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1045 contigs containing 5878070 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7070 new features were called, of which 555 are non-coding. Output genome has the following feature types: Coding gene 6515 Non-coding repeat 520 Non-coding rna 35 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.018__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 879 contigs containing 5845719 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7118 new features were called, of which 521 are non-coding. Output genome has the following feature types: Coding gene 6597 Non-coding repeat 483 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.031__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 418 contigs containing 2661803 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3081 new features were called, of which 155 are non-coding. Output genome has the following feature types: Coding gene 2926 Non-coding repeat 132 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.299__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 975 contigs containing 3813806 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5067 new features were called, of which 678 are non-coding. Output genome has the following feature types: Coding gene 4389 Non-coding repeat 657 Non-coding rna 21 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.140__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1044 contigs containing 7514401 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 8528 new features were called, of which 507 are non-coding. Output genome has the following feature types: Coding gene 8021 Non-coding repeat 473 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.500__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 460 contigs containing 2886204 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3367 new features were called, of which 277 are non-coding. Output genome has the following feature types: Coding gene 3090 Non-coding repeat 243 Non-coding rna 34 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.121__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 690 contigs containing 3098199 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3727 new features were called, of which 174 are non-coding. Output genome has the following feature types: Coding gene 3553 Non-coding repeat 150 Non-coding rna 24 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.187__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 910 contigs containing 3396558 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3948 new features were called, of which 132 are non-coding. Output genome has the following feature types: Coding gene 3816 Non-coding repeat 115 Non-coding rna 17 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.312__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2044 contigs containing 4795834 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7258 new features were called, of which 1332 are non-coding. Output genome has the following feature types: Coding gene 5926 Non-coding repeat 1320 Non-coding rna 12 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.097__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 456 contigs containing 2587168 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2956 new features were called, of which 61 are non-coding. Output genome has the following feature types: Coding gene 2895 Non-coding repeat 29 Non-coding rna 32 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.653__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 76 contigs containing 5130382 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5269 new features were called, of which 219 are non-coding. Output genome has the following feature types: Coding gene 5050 Non-coding repeat 151 Non-coding rna 68 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.828__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 286 contigs containing 2178201 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2523 new features were called, of which 115 are non-coding. Output genome has the following feature types: Coding gene 2408 Non-coding repeat 102 Non-coding rna 13 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.348__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 697 contigs containing 5586058 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6771 new features were called, of which 720 are non-coding. Output genome has the following feature types: Coding gene 6051 Non-coding repeat 687 Non-coding rna 33 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.740__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 801 contigs containing 4581472 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5568 new features were called, of which 440 are non-coding. Output genome has the following feature types: Coding gene 5128 Non-coding repeat 417 Non-coding rna 23 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.606__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1489 contigs containing 3954335 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 6182 new features were called, of which 1422 are non-coding. Output genome has the following feature types: Coding gene 4760 Non-coding repeat 1385 Non-coding rna 37 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.083__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 897 contigs containing 4309067 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4905 new features were called, of which 385 are non-coding. Output genome has the following feature types: Coding gene 4520 Non-coding repeat 365 Non-coding rna 20 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.307__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 2045 contigs containing 4438553 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 5578 new features were called, of which 264 are non-coding. Output genome has the following feature types: Coding gene 5314 Non-coding repeat 248 Non-coding rna 16 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.566__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 809 contigs containing 2689932 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3842 new features were called, of which 514 are non-coding. Output genome has the following feature types: Coding gene 3328 Non-coding repeat 495 Non-coding rna 19 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.192__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 120 contigs containing 4059739 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3927 new features were called, of which 60 are non-coding. Output genome has the following feature types: Coding gene 3867 Non-coding repeat 12 Non-coding rna 48 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.275__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 58 contigs containing 3594600 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 3586 new features were called, of which 69 are non-coding. Output genome has the following feature types: Coding gene 3517 Non-coding repeat 20 Non-coding rna 49 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. FDR.436__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1482 contigs containing 5650335 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 7668 new features were called, of which 268 are non-coding. Output genome has the following feature types: Coding gene 7400 Non-coding repeat 224 Non-coding rna 44 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.474__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 217 contigs containing 2446318 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2796 new features were called, of which 129 are non-coding. Output genome has the following feature types: Coding gene 2667 Non-coding repeat 82 Non-coding rna 47 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CLV.332__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 592 contigs containing 3337516 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4068 new features were called, of which 380 are non-coding. Output genome has the following feature types: Coding gene 3688 Non-coding repeat 350 Non-coding rna 30 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.774__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 481 contigs containing 1794449 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 2166 new features were called, of which 50 are non-coding. Output genome has the following feature types: Coding gene 2116 Non-coding repeat 10 Non-coding rna 40 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. CNT.009__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 202 contigs containing 996624 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 1345 new features were called, of which 174 are non-coding. Output genome has the following feature types: Coding gene 1171 Non-coding repeat 136 Non-coding rna 38 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.559__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1042 contigs containing 7641340 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 9873 new features were called, of which 797 are non-coding. Output genome has the following feature types: Coding gene 9076 Non-coding repeat 733 Non-coding rna 64 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.013__Fire_bins succeeded! Some RAST tools will not run unless the taxonomic domain is Archaea, Bacteria, or Virus. These tools include: call selenoproteins, call pyrroysoproteins, call crisprs, and call prophage phispy features. You may not get the results you were expecting with your current domain of U. The RAST algorithm was applied to annotating a genome sequence comprised of 1171 contigs containing 3498523 nucleotides. No initial gene calls were provided. Did not call selenoproteins because the domain is U Did not call pyrrolysoproteins because the domain is U Did not call crisprs because the domain is U Standard features were called using: glimmer3; prodigal. A scan was conducted for the following additional feature types: rRNA; tRNA; repeat regions. The genome features were functionally annotated using the following algorithm(s): Kmers V2; Kmers V1; protein similarity. In addition to the remaining original 0 coding features and 0 non-coding features, 4645 new features were called, of which 489 are non-coding. Output genome has the following feature types: Coding gene 4156 Non-coding repeat 475 Non-coding rna 14 Overall, the genes have 0 distinct functions. The genes include 0 genes with a SEED annotation ontology across 0 distinct SEED functions. The number of distinct functions can exceed the number of genes because some genes have multiple functions. KNG.177__Fire_bins succeeded!






