Edirisinghe J.N., Faria J.P., Harris N.L., Allen B.H., Henry C.S. (2018) Reconstruction and Analysis of Central Metabolism in Microbes. In: Fondi M. (eds) Metabolic Network Reconstruction and Modeling. Methods in Molecular Biology, vol 1716. Humana Press, New York, NY
DOI https://doi.org/10.1007/978-1-4939-7528-0_5
Print ISBN 978-1-4939-7527-3
Online ISBN 978-1-4939-7528-0
* Corresponding authors: JNE : ([email protected]), CSH : ([email protected])
NOTE: This tutorial is view-only, allowing you to see, but not alter, the input and output of the KBase apps used in this workflow. To run the steps yourself in a new Narrative using your own data or different parameters, copy this Narrative using the "copy" button at the top right. If you just want to read this Narrative (without copying it), you still can see the data objects generated in the workflow by using the “Controls” link at the top left. For more information, please see the Narrative Interface User Guide.
Central carbon metabolism is a key component in the metabolic network of living organisms as these pathways harbor many of the most important mechanisms for energy biosynthesis, as well as producing the precursor compounds for most essential biomass building blocks. The energy production strategies defined in the central metabolic pathways have a significant impact on the behavior and growth conditions of microorganisms, thus playing a crucial role in the quantitative prediction of biomass and energy yields [1,2]. Energy production strategies in microbes are highly diversified, unlike those in higher eukaryotes. These strategies primarily depend on environmental factors such as: (i) carbon source utilization; (ii) ability to respire by reducing numerous electron acceptors; and (iii) fermentation capabilities.
It continues to be challenging to make accurate computational predictions based on metabolic models and in silico simulations interpreting complex microbial behavior. Tools for automated metabolic model reconstruction such as ModelSEED [3-5] can rapidly generate draft genome-scale metabolic models from annotated genome sequences [6]. However, these draft models, and in some cases even curated published models, can lack accuracy in predicting growth yields, ATP production yields, and central carbon flux profiles. This poor accuracy stems primarily from three common problems: (i) poor representation of energy biosynthesis pathways; (ii) a lack of diverse electron transport chain (ETC) variations; and (iii) addition of extensive gapfilling reactions that can sometimes misrepresent an organism’s behavior [7].
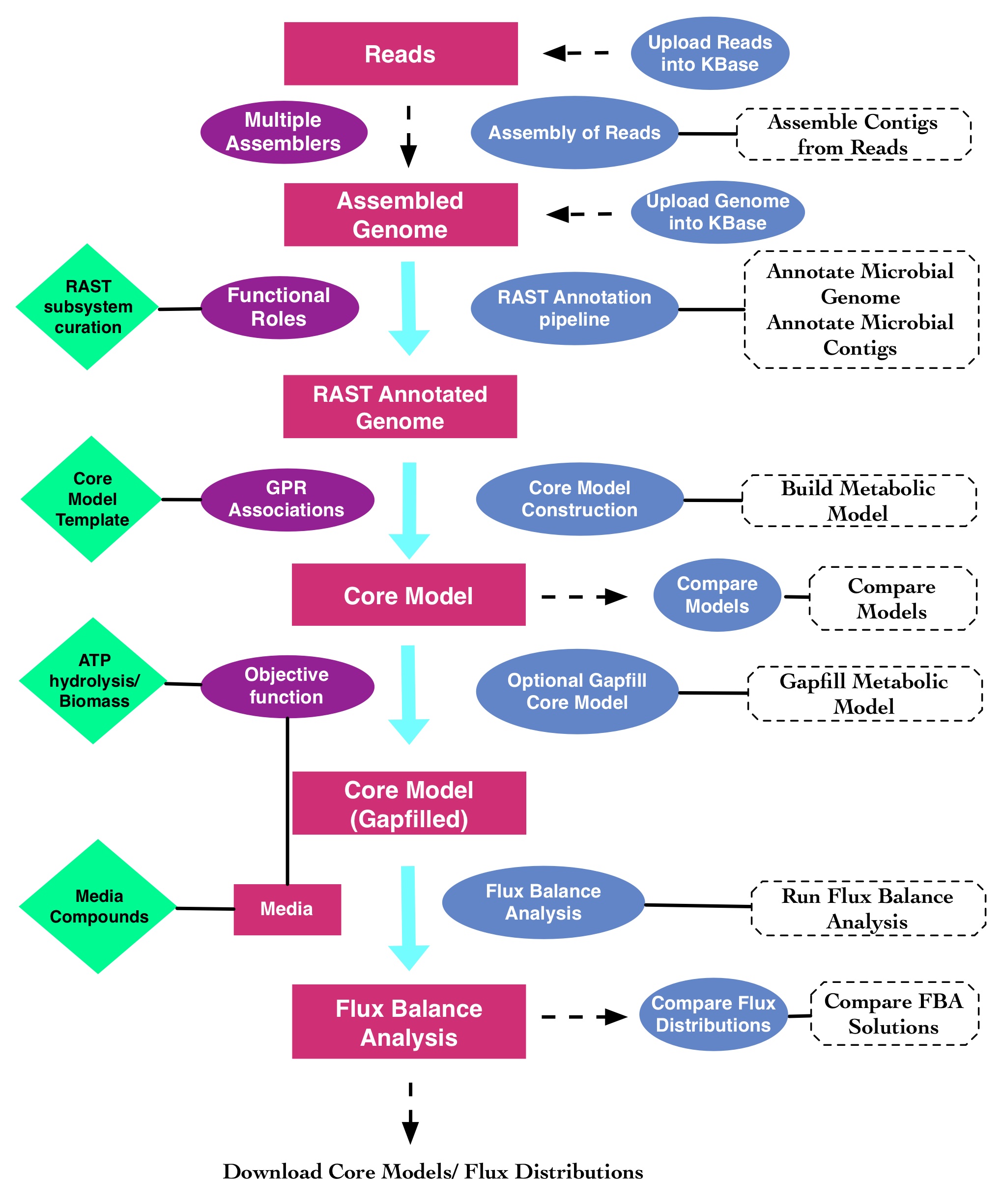
Many of these problems can be avoided by using a simplified model comprised of only the most confidently annotated and biologically critical pathways for energy biosynthesis [8] (Fig. 1). We define these models as Core Metabolic Models (CMM), and they consist primarily of the sugar oxidation pathways, the fermentation pathways (Fig. 2), and the ETC variations. We previously developed an approach for the reconstruction and analysis of CMMs based on annotated genome sequences [9], which we implemented as a pipeline in the DOE Systems Biology Knowledgebase (KBase). In this chapter, we demonstrate how this analysis workflow can be run in KBase. The complete workflow, including example data and commentary are displayed here in this Narrative. The pipeline is comprised of four main steps: (i) genome annotation by RAST [10]; (ii) CMM reconstruction [9]; (iii) gapfilling [7]; and (iv) flux balance analysis (FBA) [11]. We also discuss methods for exploring metabolic diversity by studying the variations in central metabolic pathways in a phylogenetic context.
The pipeline starts with an assembled genome with gene annotations assigned by the RAST annotation pipeline. Next, the CMMs are constructed based on a manually curated CMT that consists of GPR mappings derived from a phylogenetically diverse set of model organisms including Escherichia coli, Bacillus subtilis, Pseudomonas aeroginosa, Clostridium acetobutylicum, and Paracococcus denitrificans. As an optional step, CMMs could be gapfilled; however, most of the core models do not require any gapfilling. In the final step, FBA is performed, optimizing the biomass or ATP hydrolysis as the objective function. The pipeline also supports the comparison of the CMMs and metabolic flux distributions. Rectangles with dotted borders show the name(s) of the apps for each step.
Above, the Escherichia coli K12 genome is shown in a genome viewer. This viewer provides a concise, text-based overview of the genome as well as its contigs and genes.
In the Contigs and Genes tabs, each entry is clickable, opening either a browser for the contig or another tab with expanded information about the gene.
You can sort these entries by clicking on a column header to sort by that field (e.g., Length). Clicking the same column header again will reverse the sort order.
This Escherichia coli genome is faily complete and has a single contig: click on the contig to see neighboring genes and potential operons in this species.
To further explore this genome, click the genome name at the top of the viewer. This will open a Landing Page for the genome in a new tab in your browser. The Landing Page provides more details about the organism, its genome, and annotations.
Metabolic models generally require an objective function (OF) that is optimized during flux balance analysis to predict flux profiles. However, in our Core Metabolic Models, we explored two OFs: a biomass biosynthesis objective function and an ATP hydrolysis objective function. While CMMs do not include the amino acids, nucleotides, lipids, and cofactors that are typically included in the biomass biosynthesis objective function of genome-scale models, they do include the central carbon precursor metabolites for these compounds. Thus the biomass biosynthesis OF for our CMMs was constructed based on the biomass precursor stoichiometry derived by Varma and Parlsson and used in one of the earliest models of E. coli. When analyzing CMMs using the biomass biosynthesis OF, we found that occasionally gapfilling was required to enable synthesis of all essential biomass precursors . To permit a focused study of energy biosynthesis in our models without gapfilling, we developed a second OF for our CMMs consisting only of the ATP hydrolysis reaction: ATP + H2O -> ADP + Pi + H+. Using this OF, we computed ATP production yields in all models without any gapfilling; hence, these computations were based solely on reactions derived from existing RAST annotations
First, we will use the Build Metabolic Model app to build an initial draft metabolic core model based on the gene annotations in the Escherichia coli K12 genome. We chose Core metabolism as the model template listed under the field Template for reconstruction. This app has two steps; when the first step (Build Metabolic Model) finishes, the second step (Gapfill Metabolic Model) starts automatically.
The gapfill step lets you specify a media condition (i.e., the metabolites available in the environment in which you want to analyze your organism’s growth). If you leave the Media field blank, "complete" media will be used by default. Complete media is a special type of media that does not include an exact list of compounds. Instead, complete media consists of all metabolites for which a transporter is available in the KBase biochemistry database. (Transporters are reactions that move metabolites across cell membranes.) In the case of core models we use a minimal media for our simulations (e.g., Glucose minimimal media or Glycerol minimal media).
In addition to the media formulations available in KBase, you can upload your own custom media. In this example, Escherichia coli K12 was tested for growth in a minimal media condition called Glucose-aerobic.
We are making a preliminary assertion that a model cannot make all required biomass components from the sources in the minimal media, however, core models are desigined based on highly curated template that many of the core models inculding the one based on Escherichia coli K12 does not require any gapfilling reactions added to the model (see Table 2) in order to proudce its biomass when using Glucose minimal media as the sole carbon source.
Below, you will see the input cells for running the Build Metabolic Model app on our annotated E. coli genome.
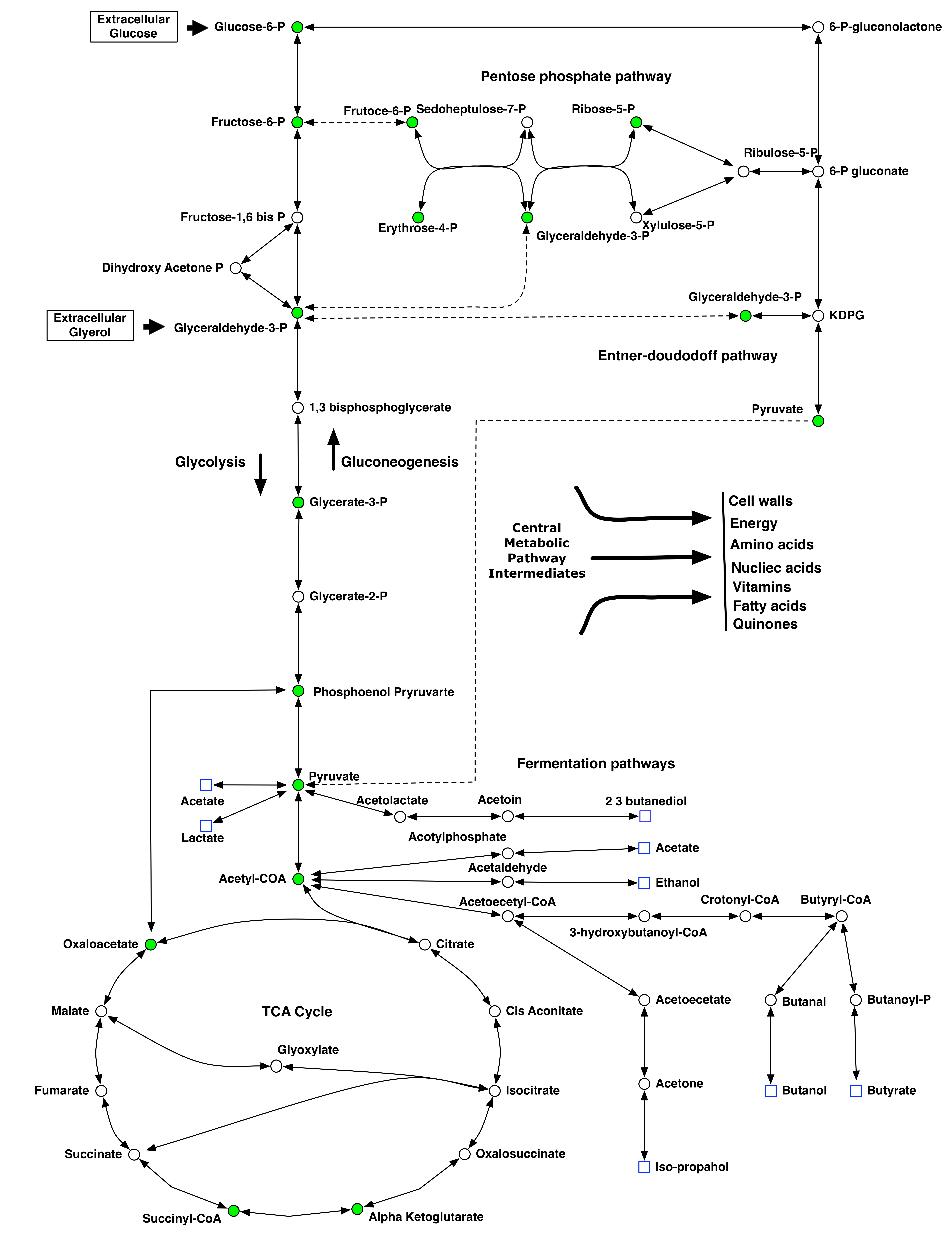
Core model pathway map displays sugar oxidation (glycolysis, gluconeogenesis, Enter-Doudoroff, pentose phosphate), TCA cycle and fermentation pathways. Central metabolic pathway metabolites produce key precursors that lead to production of all essential structural and functional components of the required for cell growth and maintenance. We have used biomass biosynthesis equation (Varma and Palsson 1993) in analyzing core metabolic model’s ability to produce these key metabolites in central metabolism; those compounds are colored in green. Fermentation pathway end products are displayed in squares with blue color borders.
An initial draft model of _Model of Escherichia coli K12 is produced based on RAST annotations. The model was not gapfilled, as the gapfilling option was not selected.
Above is the core model for Escherichia coli K12.
There are seven tabs for browsing the data in the model: Overview, Reactions, Compounds, Genes, Compartments, Biomass, Gapfilling and Pathways. The contents of these tabs are as follows:
Building Core Model of Paracoccus denitrificans PD1222
An initial draft model of _Model of Paracoccus denitrificans PD1222 is produced based on RAST annotations. The model was not gapfilled, as the gapfilling option was not selected.
Above is the core model for Paracoccous denitrificans PD1222.
There are seven tabs for browsing the data in the model: Overview, Reactions, Compounds, Genes, Compartments, Biomass, Gapfilling and Pathways. The contents of these tabs are as follows:
We have built a core metabolic model of Escherichia coli K12 ; now we can use the Run Flux Balance Analysis method to perform FBA to calculate the flow of metabolites through our model. FBA results can be used to predict the growth rate of an organism under certain conditions or the production rates for particular metabolites of interest. In this case we have used ATP hydrolysis (ATP+H2O -> ADP +Pi + H+) as the objective function.
To perform FBA, you must specify a media condition that you want to investigate using your metabolic model. In this example, we select the Glucose-aerobic minimal media, implying the organism grows on Glucose minimal media under aerobic conditions.
Flux balance analyis resutls (below) are organized into a table with six tabs: Overview, Reaction fluxes, Exchange fluxes, Genes, Biomass, and Pathways. You can see the objective value as 26.5 mmol of ATP/mmol of Glucose. We get this ATP yield as the organism undergoes oxidative phosphorylation by utilizing aerobic electron transport chains. Under the aerobic condition, glucose is fully oxidized into CO2, H2O and energy.
For more information on the Run Flux Balance Analysis method, see:
- Method tutorial
- Method details page, which includes an explanation of all parameters
- Metabolic modeling FAQ
We now run FBA on Ecoli_Glucose using Glucose minimal media under anaerobic conditions (without the presense of oxygen). We select the Glucose-anaerobic media formulation.
Notice the objective value is now 2.75 mmol of ATP/mmol of glucose when simulated under the anaerobic condition. Compared to the objective value 26.5 mmol of ATP/mmol of glucose under the aerobic condition, it is significantly less. This is because there is no oxygen present in the media. As a result, oxidative phosphorylation is not active, electron transport chains are not utilized to produce energy. Under this condition, the organism produces energy solely from the fermentation process.
Facultative anerobic organisms like Escherichia coli can grow both in aerobic and anaerobic conditons, as shown before. They are able to reduce a number of anaerobic electron acceptors such as nitrate (NO3), dimethyl solfuxide (DMSO) and trimethyl amineoxide (TMAO) during anerobic respiration. If anaerobic electron acceptors are not present in the medium, these organisms are still able to grow solely using the fermentation process (as shown above). Now we run FBA on our model 'Ecoli_Glucose' anerobically with nitrate (NO3) present as an anaerobic electron acceptor.
Paracoccous denitrificans PD1222 can grow both in aerobic and anaerobic conditons, as shown before. They are able to reduce a number of anaerobic electron acceptors such as nitrate (NO3), dimethyl solfuxide (DMSO) and trimethyl amineoxide (TMAO) during anerobic respiration. If anaerobic electron acceptors are not present in the medium, these organisms are still able to grow solely using the fermentation process (as shown above). Now we run FBA on our model 'Ecoli_Glucose' anerobically with nitrate (NO3) present as an anaerobic electron acceptor.
Unlike the electron transport chains of higher eukaryotes, bacterial ETCs are highly diversified. As a result, they are able to grow in a variety of aerobic and anaerobic environments reducing anaerobic electron acceptors such as nitrate, nitrite, fumarate, dimethyl sulfoxide(DMSO) and trimethylamine N-oxide (TMAO). For instance, Escherichia coli (below) can respire aerobically and anaerobically reducing nitrate, fumarate, TMAO and DMSO. Paracoccus denitrificans (below) is also able to grow aerobically and able to reduce multiple nitrogen based compounds anaerobically including nitrate, nitrite, nitrous oxide and nitric oxide. Better annotation of ETCs helps us identify complex respiration types and make accurate energy yield predictions. In our CMMs, we have focused on adding these diverse ETC reactions across the bacterial tree of life that are derived from consistently assigned gene annotations.
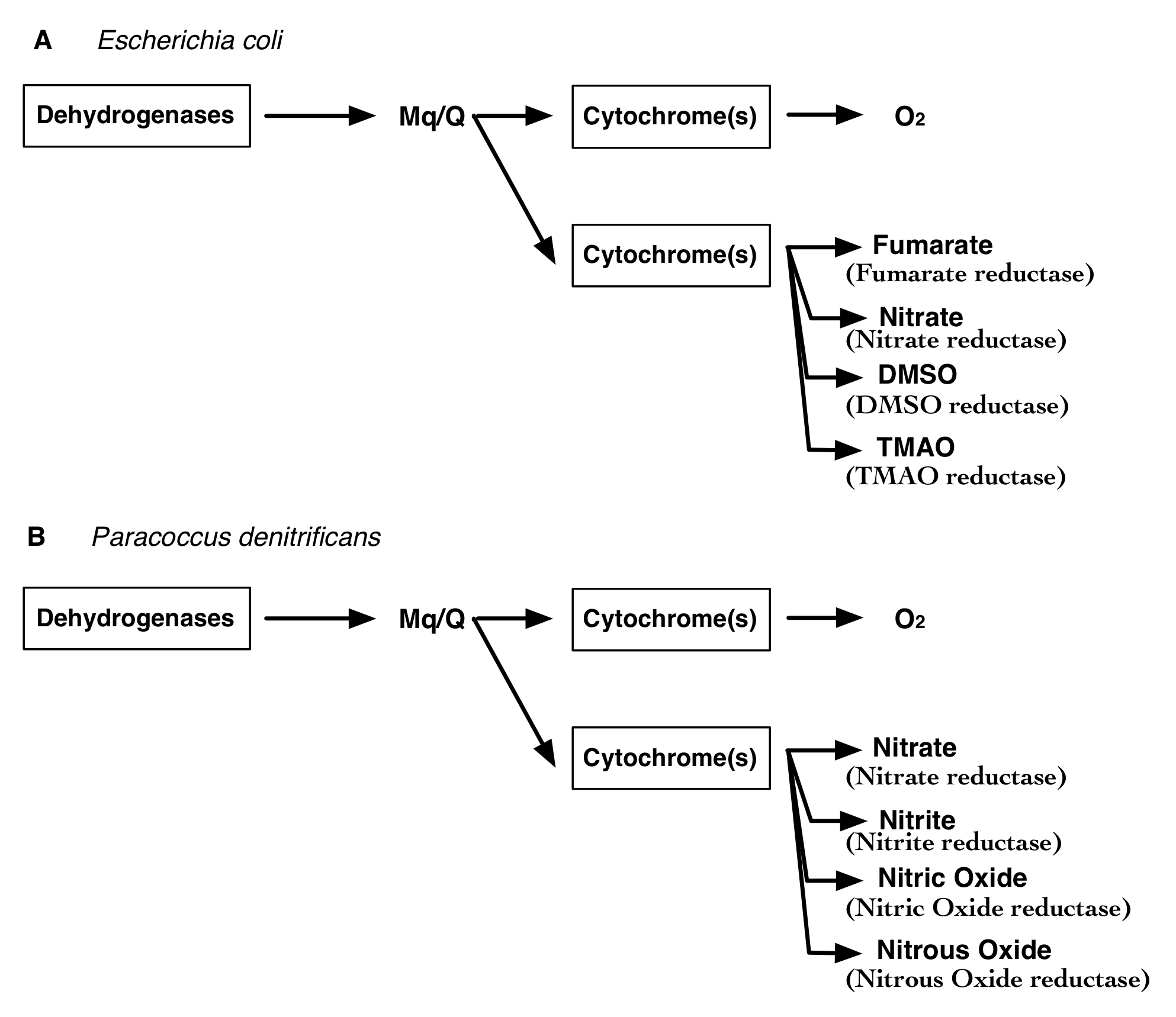
We are able to compare the two models of Escherichia coli and Paracoccous denitrificans
As seen under Flux Balance Analysis tab -> Objective column, we can see aerobic growth yield 26.5 ATP verses anaerobic growth (without any electron acceptors) yield is much lower at 2.75. You can find ATP yield data on number of model organisms here https://narrative.kbase.us/narrative/ws.15253.obj.1#ATP-yield-predictions-of-core-models-under-aerobic-and-anerobic-conditions
As explained earlier, Core Models have two objective functions, ATP hydrolysis and biomass biosynthesis. About 41% of core models (3415), including the core model of Escherichia coli, do not need any gapfilling reactions addedd in order to produce the essential biomass precursors in the OF. However, some core models (see Table 1) require gapfilling reactions to be added in order to satisfy the biomass objective function. Now we run FBA, selecting biomass (bio2) as the objective function against glucose minimal media without the gapfilling option.
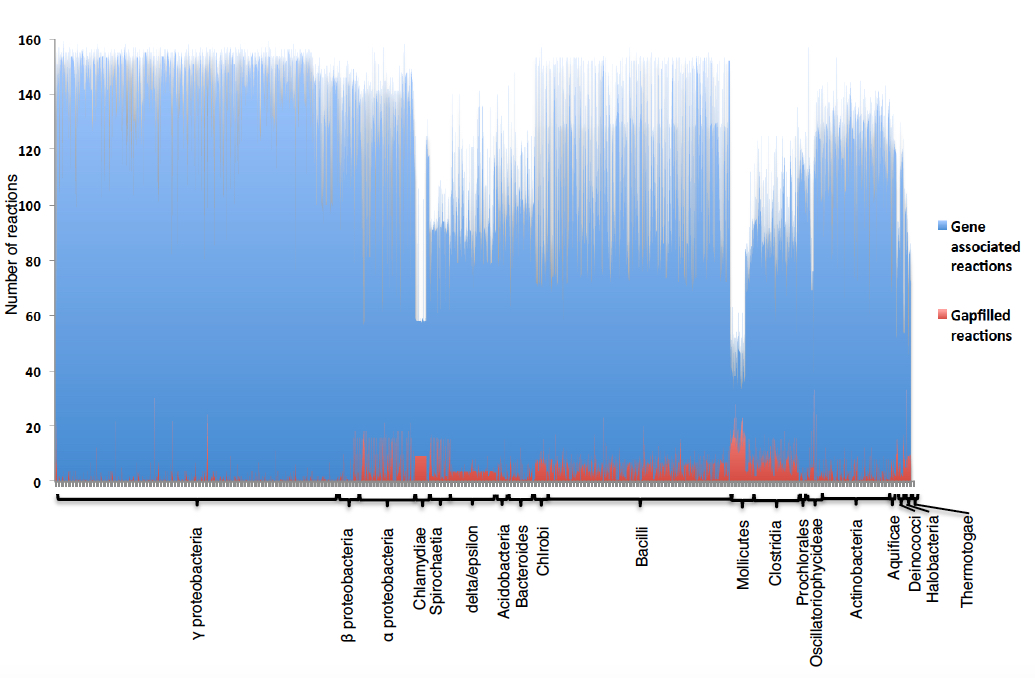
We can see the Escherichia coli core model was able to grow (0.12 objective value) without any gapfilling reactions added to the model. However, as some models do require gapfilling in order to produce biomass precursors, we have run an analysis identifying the distribution of number of gapfilling reactions needed by each model and organized them by phylogeny.
Figure 3. Number of gapfilled reactions that are required in CMMs in order to produce all biomass precursors, with CMMS organized by phylogenetic group. The blue bars represent the gene-associated reactions and the red bars represent the gapfilled reactions for all CMMs used in this study. The height of the bars represents the number of reactions. CMMs are grouped according to taxonomy.