This Narrative is part of a collection used in a single publication. The paper took a metagnomic and metatranscriptomic approach to examine a bioreactor's response to the presence of sulfate, which reduces the production of methane, a valuable product, while producing hydrogen sulfide, a corrosive contaminant.
This Narrative collection has been created to highlight the types of analyses done for this publication. They are as follows (in order):
Please cite: Andrew R St. James, Ruth E Richardson. Ecogenomics reveals community interactions in a long-term methanogenic bioreactor and a rapid switch to sulfate-reducing conditions, FEMS Microbiology Ecology, Volume 96, Issue 5, May 2020, https://doi.org/10.1093/femsec/fiaa050.
This Narrative contains the final MAGs that were published in the paper.
This publication involved a numerous layers of analyses and, as a result, elements are spread across many Narratives, some of which also contain exploratory analysis that was not continued further. Below, the bioinformatic methods used in the paper are described in detail, with links to two example Narratives, one containing a metagenomic analysis of the SJ1 samples and another containing metatranscriptomic analysis of the same sample.
| MAG | Organism name | Size (Mbp) | GC content | Contigs | Completeness (contamination) |
RRA SJ1 | RRA SJ1S | Range of RRA control MTs |
Range of RRA sulfate MTs |
|---|---|---|---|---|---|---|---|---|---|
| MAG006 | Desolfovibrionaceae sp. SJ1006 | 3.9 | 0.651 | 32 | 100.00% (0.00%) | 0.0214 | 0.2599 | 1.52–8.24% | 3.05–10.74% |
| MAG002 | Syntrophomonas sp. UBA5314 | 4.3 | 0.493 | 68 | 97.96% (3.95%) | 0.0742 | 0.0295 | 33.73–38.90% | 31.62–39.68% |
| MAG015 | Syntrophomonas sp. SJ1015 | 3.2 | 0.493 | 85 | 90.03% (2.93%) | 0.0135 | 0.0201 | 9.53–12.80% | 11.16–14.03% |
| MAG004 | Methanosaeta concilli | 2.7 | 0.519 | 94 | 100.00% (0.65%) | 0.0311 | 0.036 | 7.03–23.91% | 11.82–24.44% |
| MAG010 | Methanospirillum hungatei | 3.4 | 0.454 | 50 | 99.02% (0.98%) | 0.013 | 0.0 | 4.63–12.64% | 4.84–9.91% |
This work investigates a model butyrate-to-methane bioreactor over 48 hours of pulse feeding with butyrate and sulfate, and analyzing the metabolic flexibilty of the sulfate-reducing bacteria and their relationship to methanogenic bacteria.
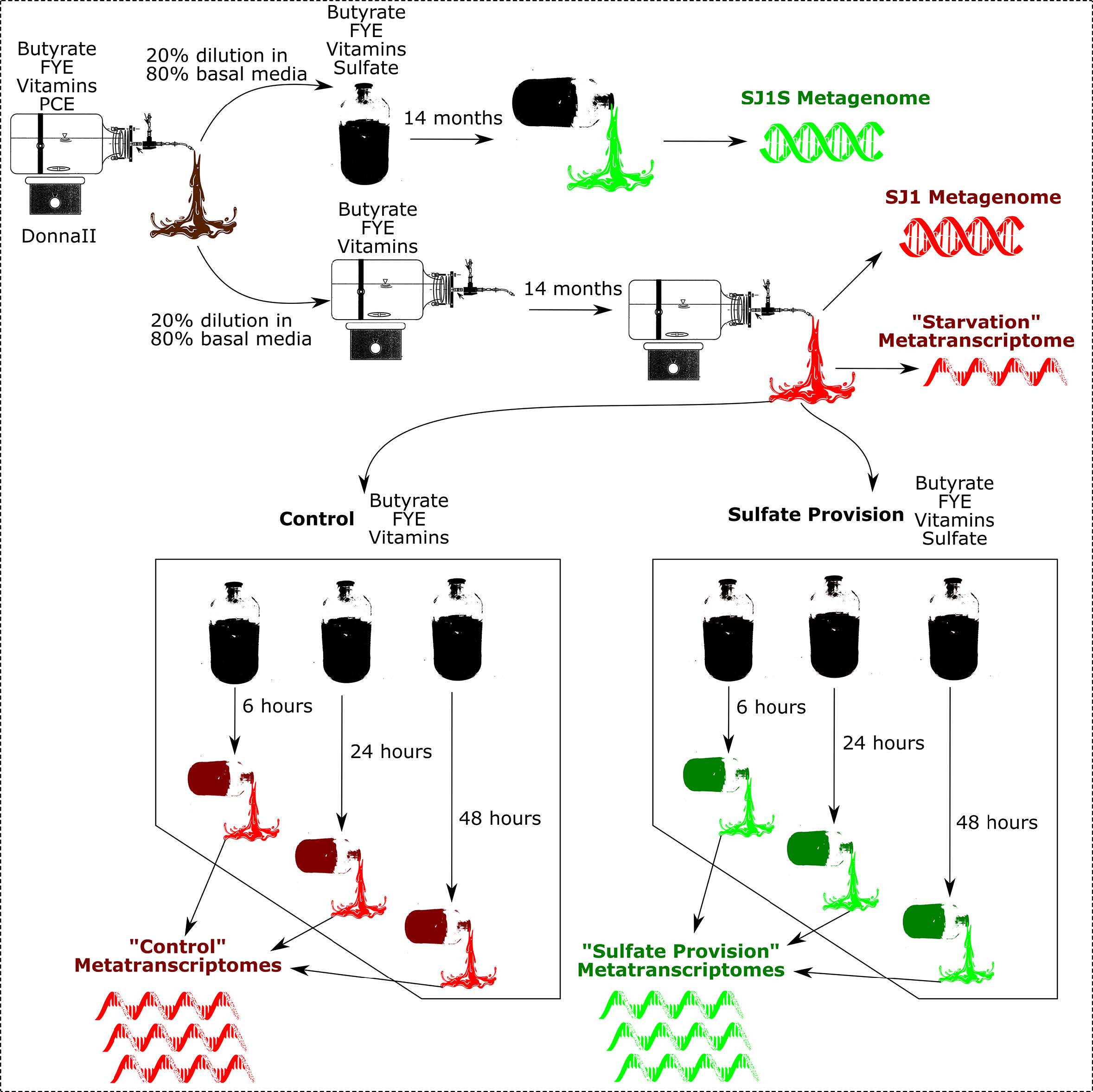 Figure 1: Overview of Sampling Scheme
Figure 1: Overview of Sampling SchemeMetatranscriptome analysis can be found in this Narrative.
QC-filtered reads were aligned using Bowtie2 (view in Narrative), and then relative abundance calculated using Cufflinks (view in Narrative).
Note: both Narrative contain exploratory analysis, and not all sections of the Narrative were included in the paper.
Proteobacteria are dominated by populations implicated in internal sulfur cycling including Sulfuricurvum as well as Desulfovirionales and Syntrophobacterales. Neither Firmicutes nor Bacteroidetes are dominated by single genera in the metagenomes. Euryarchaeota are dominated by reads mapping to the aceticlastic genus Methanosaeta and hydrogenotrophic genus Methanospirillum. In SJ1S, Proteobacteria, especially Desulfovibrionales increase while Methanospirillum and Sulfuricurvum effecively reach extinction. Binning from SJ1 resulted in 61 high-quality MAGs (>90% complete and <10% contaminated). Six of those MAGs of interest are stored in the KBase narrative cited in the paper. The MAG identified as Desulfovibrionacea sp. SJ1006 was found to show change in expression due to presence of sulfate, indicating the MAG shifts its physiological strategy in response to sulfate. The MAG identified as Syntrophomonas sp. SJ1015 was found in increase central carbon metabolism in response to sulfate.
For full results and discussion, please see the paper.