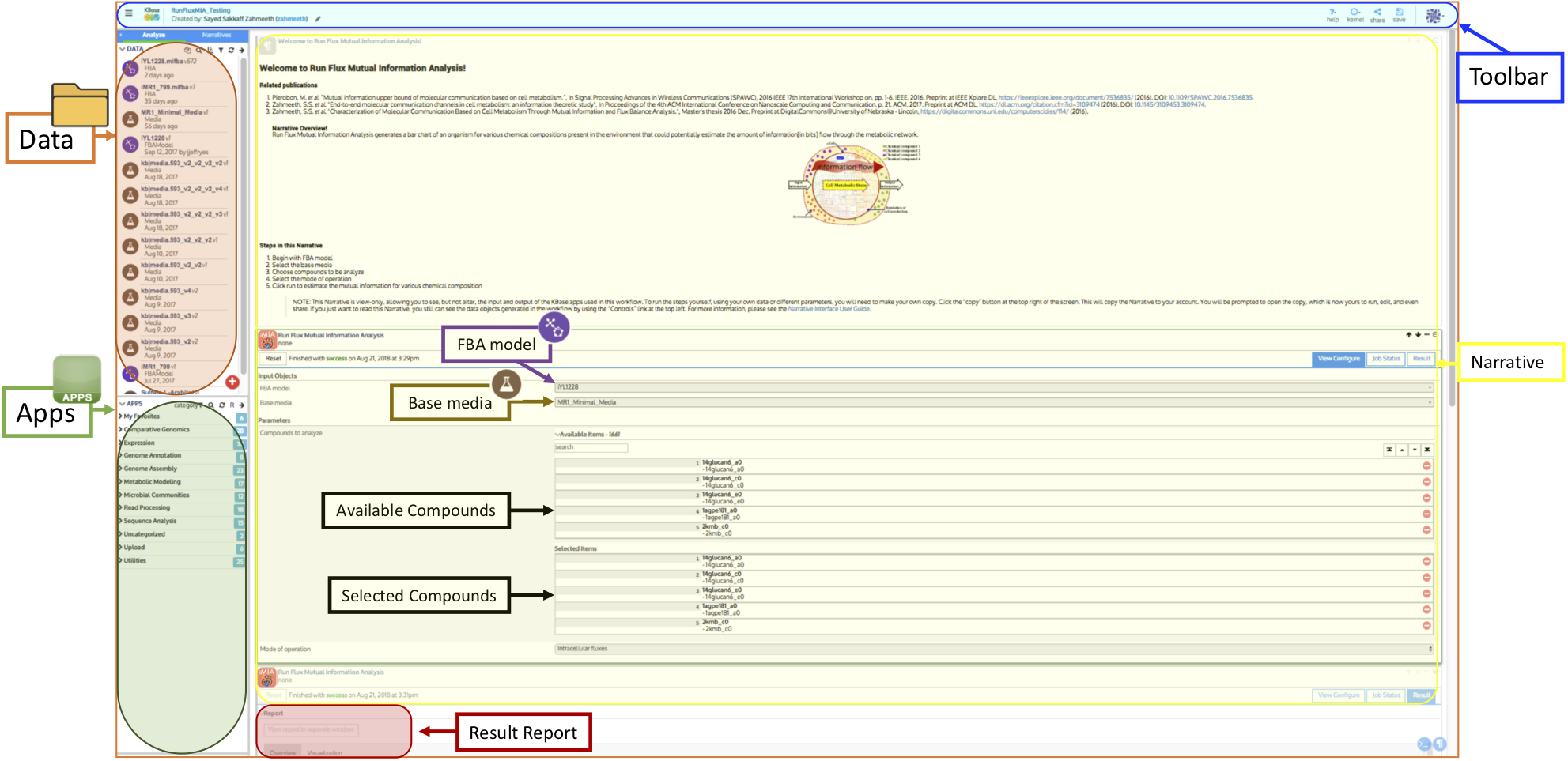
Explore the mutual information between model flux and media inputs
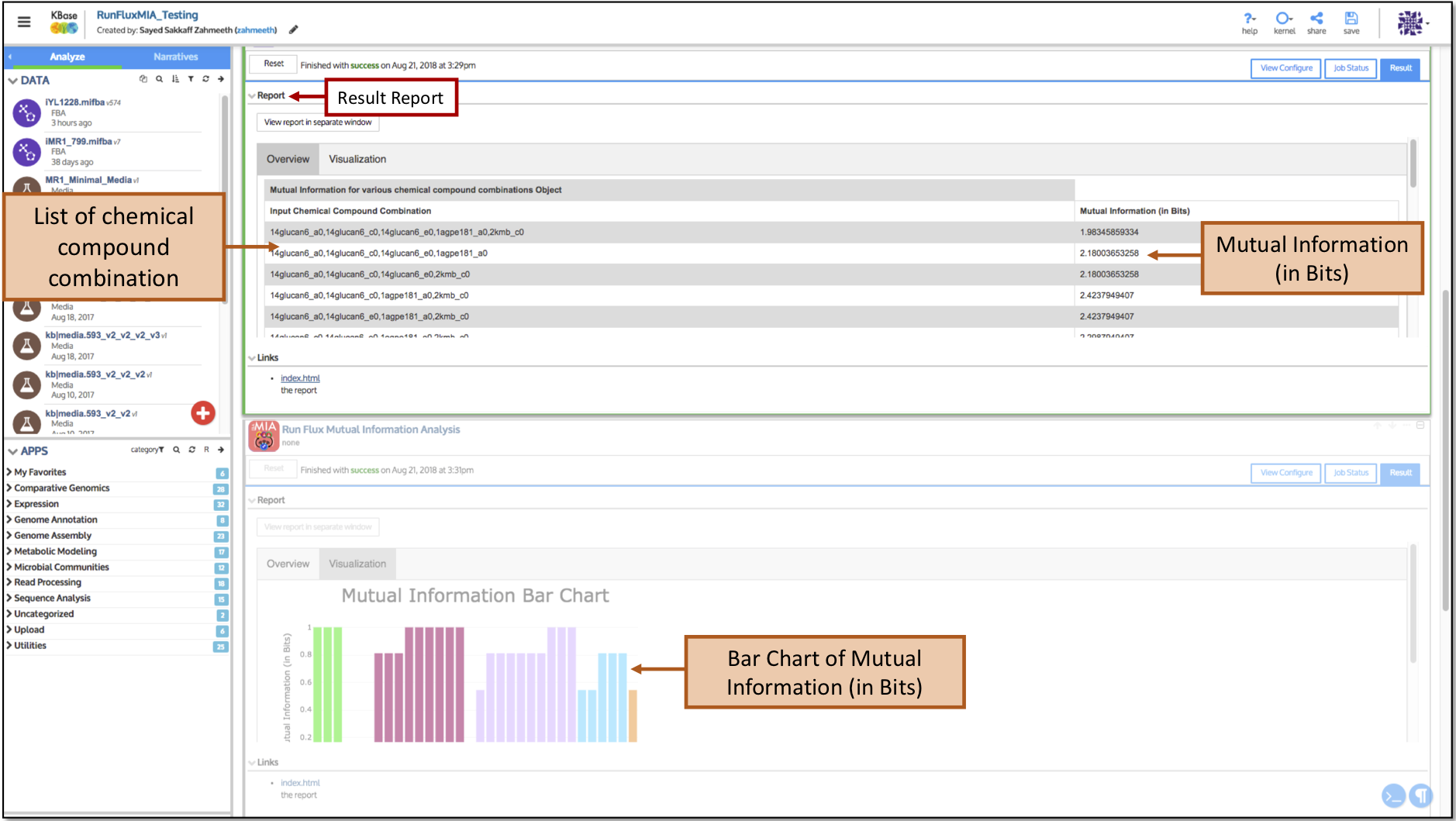
Goal: This app generates a bar chart of an organism for various chemical compositions present in the environment that could potentially estimate the amount of information in bits flow through the metabolic network.
We demonstrate that this study can lead to a ranking of chemical compounds on the basis of how much mutual information through the through the metabolic network can result from their variation.
This can potentially lead to the reduction in the number of expensive experiments by reducing the different combinations of chemical compounds necessary to obtain and study determinate metabolic behaviors of these organisms, which might be associated with dysbiosis.
Begin by selecting the metabolic FBA model of an organism that you want to perform the Run Flux Mutual Information Analysis upon. Then specify the base media on which you will grow your organism. Finally, select the mode of operation such as mutual information from intracellular flux values; biomass production; or section, uptake, and biomass production
Team members who developed & deployed the mathematical models & deployed algorithms in KBase: Zahmeeth Sakkaff, Massimilliano Pierobon and Chris Henry, For questions, contact us.
Acknowledgments - We would like to thank the KBase team for their support in development, special thanks to: James Jeffryes, Tian Gu, Filipe Liu, Boris and Janaka Edirisinghe.
Related Publications
- [1] Pierobon, M. et al. Mutual information upper bound of molecular communication based on cell metabolism. , In Signal Processing Advances in Wireless Communications (SPAWC), 2016 IEEE 17th International Workshop on, pp. 1-6. IEEE, 2016. Preprint at IEEE Xplore DL. , https://ieeexplore.ieee.org/document/7536835/
- [2] Zahmeeth, S.S. et al. "End-to-end molecular communication channels in cell metabolism: an information theoretic study", In Proceedings of the 4th ACM International Conference on Nanoscale Computing and Communication, p. 21. ACM, 2017. Preprint at ACM DL. , https://dl.acm.org/citation.cfm?id=3109474
- [3] Zahmeeth, S.S. et al. "Characterization of Molecular Communication Based on Cell Metabolism Through Mutual Information and Flux Balance Analysis.", Master's thesis 2016 Dec. Preprint at DigitalCommons@University of Nebraska - Lincoln. , https://digitalcommons.unl.edu/computerscidiss/114/
App Specification:
https://github.com/zahmeeth/MutualInformationAnalysisModule/tree/4a36f36af2ab8cff4e54c9e7fc6dac1955bb5037/ui/narrative/methods/run_flux_mutual_information_analysisModule Commit: 4a36f36af2ab8cff4e54c9e7fc6dac1955bb5037